Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> Insights > SARS-CoV-2 Spike protein mutation may increase the virus infectivity The Spike protein of SARS-CoV-2 facilitates the viral entry into target cell exploiting Angiotensin-converting enzyme 2 (ACE2) receptor. [1][2] The Spike protein receptor-binding domain (RBD), which is on the S1 subunit of SARS-CoV-2, is the critical determinant of viral tropism and infectivity. The genome analysis of global SARS-CoV-2 strains yields 32 RBD mutant strains clustering into 10 mutation types under high positive selection pressure. Three mutation types circulating in Wuhan, Shenzhen, Hong Kong, and France, displayed enhanced structural stability along with higher human ACE2 receptor affinity of the spike protein, indicating these mutants may have acquired increased infectivity to humans. [3]
SARS-CoV-2 RBD mutation mapping
It is believed that the RBD should be highly conserved because it is the only domain to bind human ACE2 and initiate cell entry. Surprisingly, the RBD sequences were as diverse as the other regions of the S protein, revealed by polymorphism and divergence analysis.
Among the 1609 SARS-CoV-2 strains in the public database, 32 strains contained amino acid mutations in the RBD. These mutant strains were reported from multiple locations including China, the UK, Finland, France, Belgium, the USA, and India, most of which had only one amino acid change compared to the originally reported genome.
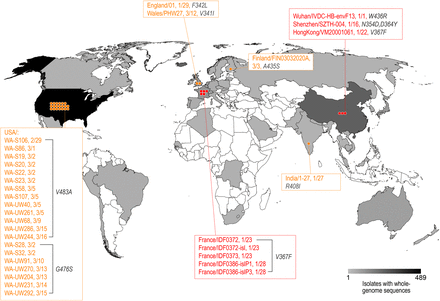
Fig. 1: Distribution of the SARS-CoV-2 strains mutated in the RBD of the S protein. The geographic distribution of the RBD mutant strains is displayed. The strains with names highlighted in red are mutants with the enhanced binding affinity. The strains with names noted in yellow are mutants with similar binding affinities.
(Source: bioRxiv preprint doi: https://doi.org/10.1101/2020.03.15.991844)
Three Mutation types bind human ACE2 receptor with higher affinityffinity
Molecular dynamics simulations were performed to estimate the functional change suggested by the RBD mutations. Three RBD mutant types (N354D and D364Y, V367F, W436R) exhibited significantly lowered binding free energy (ΔG), suggesting a significantly increased affinity to human ACE2. [3]
Structural basis for increased affinity
The binding surface of the RBD to ACE2 is largely in random coil conformation, which lacks structural rigidity. Therefore, a firm scaffold should help to maintain this conformation of the interaction surface, and thus may facilitate the binding affinity. The beta-sheet structure scaffold, centered by residues 510-524 (Fig. 2, marked as red), provides this rigidity.
“Higher affinity” mutants (N354D D364Y, V367F, and W436R) exhibited a general decreased ΔG in the binding site region, which increased the structural stability. What’s more, the mutation W436R provides a positively charged Arg in the proximity of the complementing highly negative charged ACE2 surface. This potential electrostatic attraction may contribute to the higher affinity.[3]
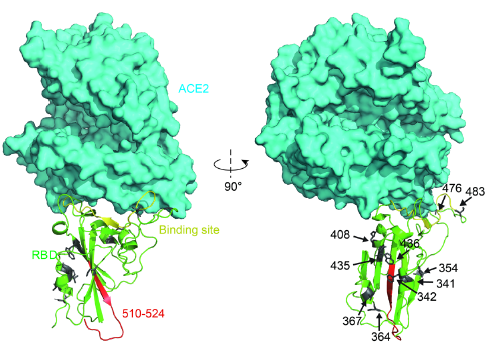
Fig. 2: Structural analysis of RBD mutants and the effects on their binding affinity. Spatial 409 location of the mutant amino acids and the fragment 510-524.
(Source: bioRxiv preprint doi: https://doi.org/10.1101/2020.03.15.991844)
Reference:
1. Hoffmann et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, 2020, Cell 181, 271–280;
2. Zhou et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, 2020, Nature 579, 270-273;
3. Ou et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein, bioRxiv preprint doi: https://doi.org/10.1101/2020.03.15.991844;
To investigate the findings discussed above, ACROBiosystems expressed the recombinant prototype spike protein RBD and “higher affinity” mutants in house. They used three different methods, including ELISA, Biacore SPR, and Octet BLI, to analyze the binding affinities between human ACE2 protein and different spike proteins respectively. The results were summarized in Fig. 3, Fig. 4, and Fig. 5 below. As you can see, all the “higher affinity” spike protein RBD mutants showed stronger binding affinities compared to the prototype.
Bioactivity-BLI
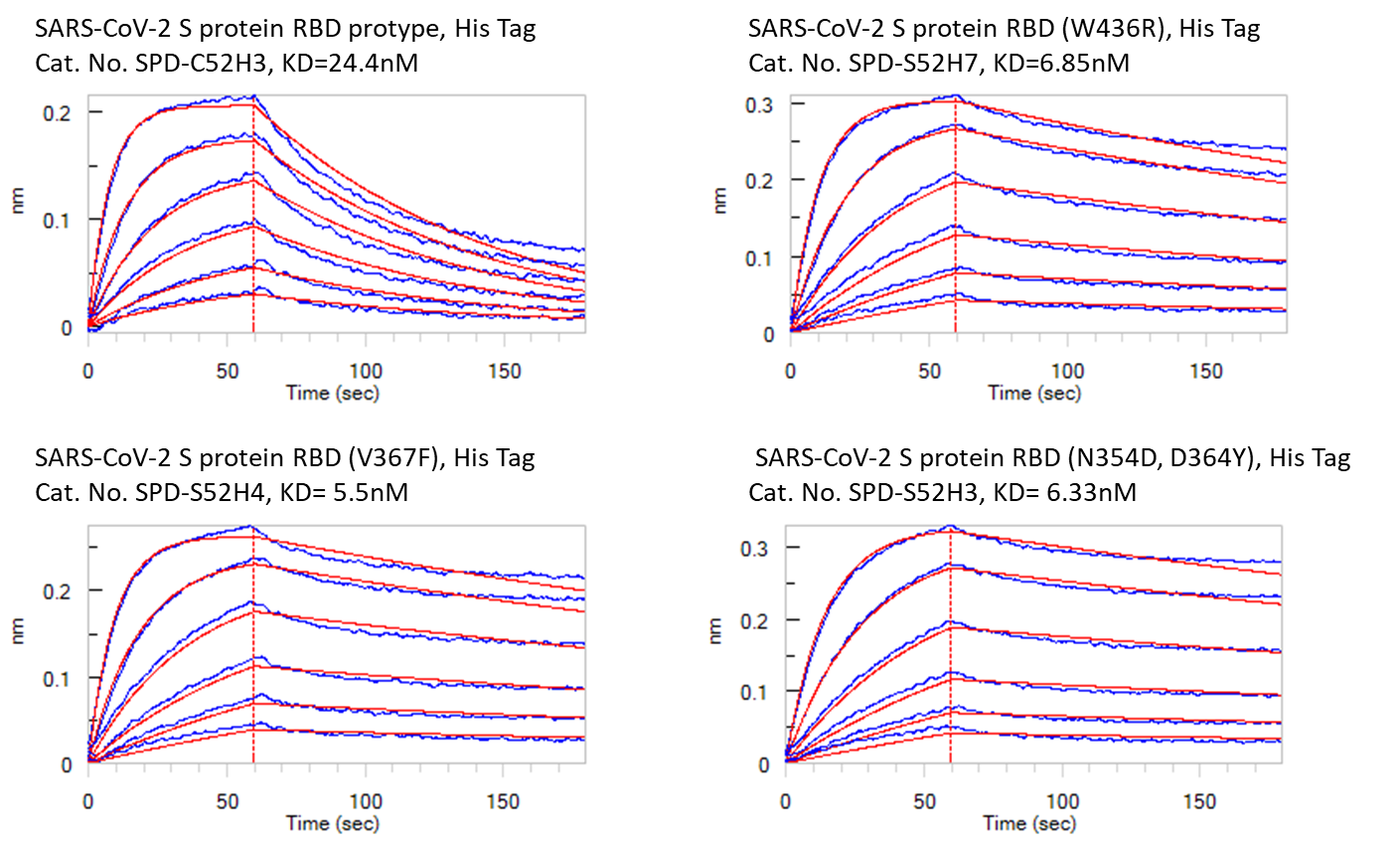
Fig. 4: Bioactivity – Octet BLI results. The KD between Human ACE2 and prototype spike protein RBD is 24.4nM. The KD between Human ACE2 and RBD(V367F), RBD (W436R), and RBD (N354D, D364Y) were 5.5nM, 6.85nM, and 6.33nM respectively.
This web search service is supported by Google Inc.







