 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> resDetect™ Therapeutic Antibody & Recombinant Proteins CMC Process Residual Quality Control Solutions

Browse all our resDetect™ products
Therapeutic antibodies & recombinant proteins are important critical biopharmaceuticals used to treat variety of diseases. However, their complex production processes can generate various impurities, particularly CMC process residues such as host cell DNA and Protein A residues. These residual impurities can impact the pharmacokinetics, pharmacodynamics, and immunogenicity of the drugs, potentially causing adverse reactions and treatment failures. Ensuring the safety, efficacy, and purity of these drugs is essential and strict regulatory requirements mandate the control of residual impurities in antibody and recombinant protein drugs to ensure patient safety and health.
Our brand, resDetect™, offers comprehensive quality control solutions for detecting process residues, supporting your CMC quality control studies for antibody and recombinant protein drugs.
 Stable and Reliable — Stringent batch testing and release, with inter-batch differences less than 15%.
Stable and Reliable — Stringent batch testing and release, with inter-batch differences less than 15%. Regulatory Compliance — Complaint with standards and regulations of international regulatory authorities.
Regulatory Compliance — Complaint with standards and regulations of international regulatory authorities. Global Supply— Product delivery in 1-3 days, worldwide.
Global Supply— Product delivery in 1-3 days, worldwide. Thorough Validation — Referencing guidelines such as ICH Q2 (R2) and ChP 9101.
Thorough Validation — Referencing guidelines such as ICH Q2 (R2) and ChP 9101.Host Cell Nucleic Acid Residual Detection
Affinity Ligand Residual Detection
CHO Host Cell Nucleic Acid
Residual Detection
Pichia pastoris Nucleic Acid
Residual Detection
E. coli Nucleic Acid Residual
Detection
HEK293 Host Cell Nucleic Acid
Residual Detection
CHO cells are essential in developing and producing therapeutic antibodies, including monoclonal antibodies, bispecific antibodies (bsAbs), and antibody-drug conjugates (ADCs). Their high expression capability and stability make them crucial in biopharmaceuticals. However, the production processes can leave residual host cell DNA (resDNA), which poses safety risks such as carcinogenicity and infections. Detecting resDNA is crucial for ensuring the safety and efficacyof drugs in Chemistry, Manufacturing, and Controls (CMC) quality control.
Our resDetect™ CHO resDNA Quantitation Kit (qPCR) addresses the quality control requirements for detecting CHO cell resDNA in drug CMC production.
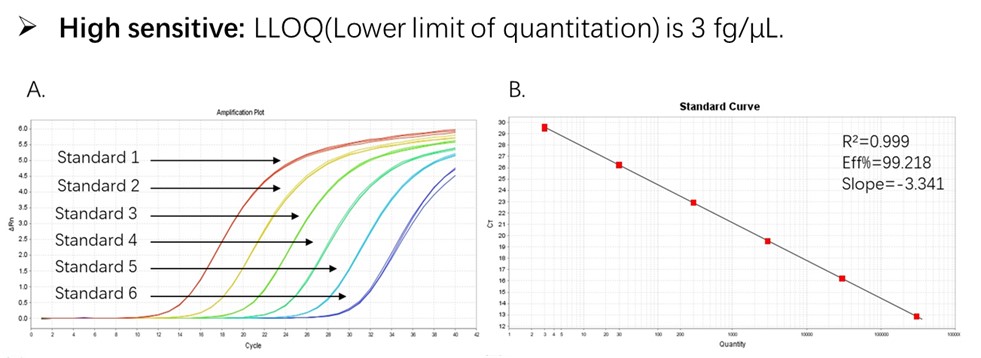
Figure 1. High sensitivity and broad dynamic range using the CHO resDNA Quantitation Kit. (A) Typical analysis results obtained with Standard 1 (300 pg/μL) to 6 (3 fg/μL). (B) The standard curve of the 10-fold dilution series. PCR efficiency should be 90-110%.
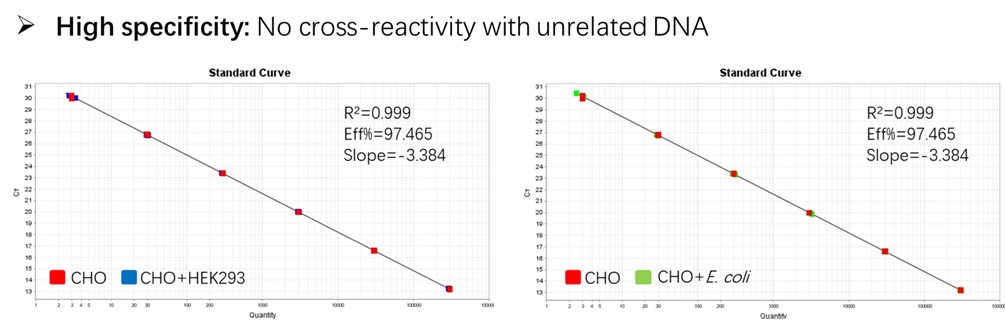
Figure 2. Assay specificity. Standard curves generated using 10-fold serial dilution of CHO genomic DNA (included in the kit).
 Resources
Resources Video —— Demonstration of CHO Host Cell Residual Detection Kit Operation
Video —— Demonstration of CHO Host Cell Residual Detection Kit Operation
 Protocol —— CHO resDNA Quantitation Kit (qPCR)
Protocol —— CHO resDNA Quantitation Kit (qPCR)
 Brochure —— resDetect™ CMC Process Residual Detection Solutions
Brochure —— resDetect™ CMC Process Residual Detection Solutions
 FAQ
FAQThe resDetect™ resDNA Sample Preparation Kit (Magnetic Beads), catalog # OPA-R005, is the ideal choice. Its elution buffer is compatible with the qPCR detection system's buffer, ensuring optimal amplification efficiency and sample recovery. Additionally, OPA-R023 is a pre-packed version of OPA-R005, designed for use with our nucleic acid pre-processing machines, saving valuable time for our customers.
Absolutely! Our sample pre-processing kits are optimized to be compatible with all major nucleic acid extraction instruments on the market. Their performance and recovery rates have been thoroughly tested and validated.
Absolutely! Our kit is robust enough to handle pH matrices ranging from 3-10 and highly concentrated protein samples up to 150 mg/mL. Rigorous in-house testing and real-world customer trials have confirmed its efficacy. No need for complex sample preparation like pH adjustments or dilutions – just plug and play!
Buffer NT and Buffer LA should be kept separate to avoid salt precipitation and a decrease in extraction efficiency. However, Buffer NT can be mixed with Proteinase K in advance to streamline the workflow in the lab!
Pichia pastoris is a highly effective expression system known for its high-density fermentation and robust expression of recombinant proteins. It offers eukaryotic post-translational modifications such as glycosylation, disulfide bond formation, and proper protein folding, making itwidely used in production of recombinant protein drugs (e.g., growth hormones, human albumin, collagen).
To support the quality control of Pichia pastoris expression systems, we offer the resDetect™ Pichia pastoris Host Residual DNA Quantitation Kit (qPCR) (Catalog number: OPA-R017). This kit utilizes quantitative PCR to precisely quantify residual host DNA (resDNA) in process samples or final products.

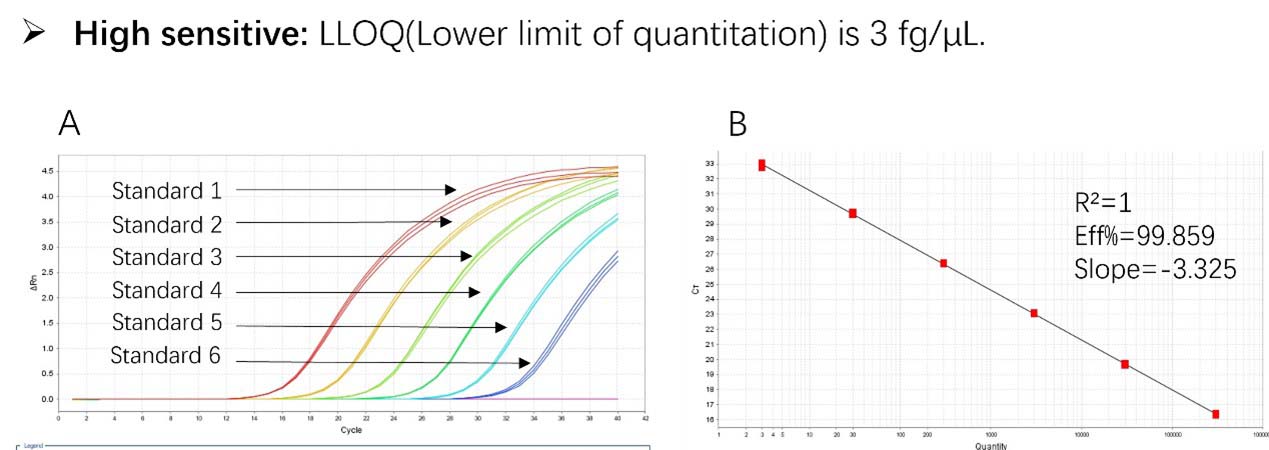
Figure 1. High sensitivity and broad dynamic range using the Pichia pastoris resDNA Quantitation Kit. (A) Typical analysis results obtained with Standard 1 (300 pg/μL) to 6 (3 fg/μL). (B) The standard curve of the 10-fold dilution series. PCR efficiency should be 90-110%.
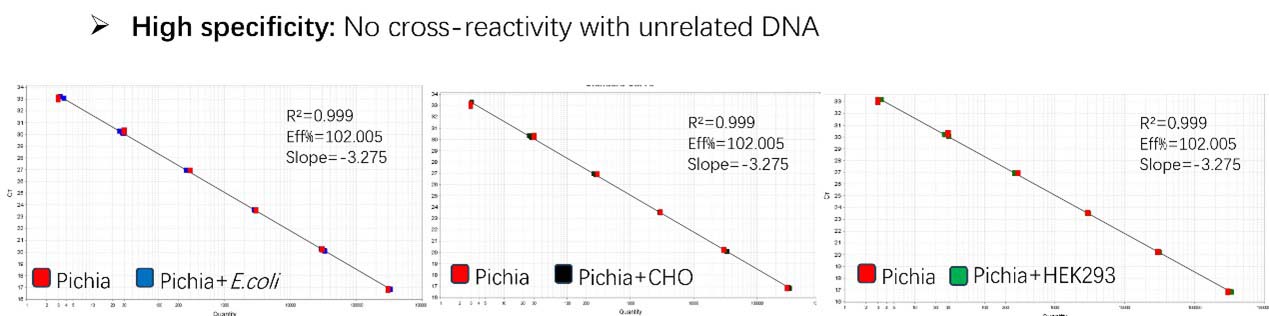
Figure 2. Assay specificity. Standard curves generated using 10-fold serial dilution of Pichia pastoris genomic DNA (included in the kit) in the presence of DNA (CHO, E. coli and HEK293).
 Resources
Resources Protocol —— resDetect™ Pichia pastoris resDNA Quantiation Kit (qPCR)
Protocol —— resDetect™ Pichia pastoris resDNA Quantiation Kit (qPCR)
 Brochure —— resDetect™ CMC Process Residual Detection Solutions
Brochure —— resDetect™ CMC Process Residual Detection Solutions
 FAQ
FAQLook no further than OPA-R024, the resDetect™ resDNA Sample Preparation Kit II (Magnetic Beads). Its elution buffer is perfectly aligned with the qPCR detection system's buffer, ensuring optimal sample amplification efficiency, recovery rates, and differentiation of NTC.
Absolutely! It is fully compatible with challenging conditions, including pH 3-10 matrix intermediates or highly concentrated protein samples (up to 150 mg/mL). Our kit has been extensively tested internally and validated with numerous customer samples, the kit simplifies extraction without requiring pre-treatment like pH adjustments or dilutions.
Buffer NT and Buffer LA should keep separate to avoid salt precipitation and maintain extraction efficiency. However, Buffer NT can be safely premixed with Proteinase K to streamline laboratory workflows.
Escherichia coli (E. coli) is widely utilized expression system for producing recombinant protein drugs because its efficient protein expression, cost-effectiveness, and rapid production cycle. However, the presence of residual E. coli host cell DNA (HCD) in final product poses potential health risks, including potential immunogenicity and adverse reactions. As a result,, stringent quality control measures are essential to ensure the safety and efficacy of biopharmaceuticals produced using the E. coli system.
Our resDetect™ E. coli resDNA Quantitative Kit (qPCR), offer a highly sensitive and specific solution for the detection and quantification of residual E. coli host cell DNA.

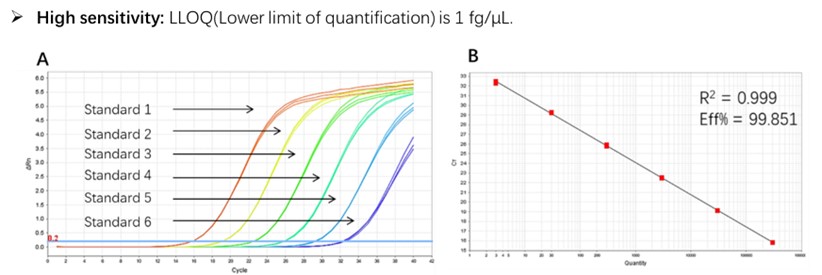
Figure 1. High sensitivity and broad dynamic range using the E. coli residual DNA quantitative Kit. (A) Typical analysis results obtained with Standard 1 (300 pg/μL) to 6 (3 fg/μL). (B) The standard curve of the 10-fold dilution series. PCR efficiency should be 90-110%.
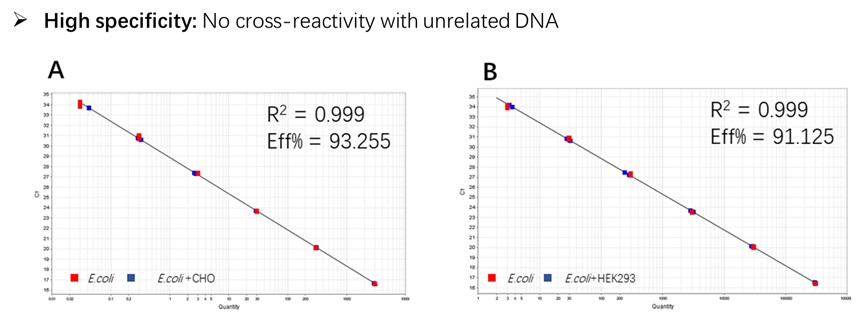
Figure 2. Assay specificity. Standard curves generated using 10-fold serial dilution of E. coli genomic DNA (included in the kit).
 Products
Products resDetect™ E. coli resDNA Quantitative Kit (qPCR)
resDetect™ E. coli resDNA Quantitative Kit (qPCR)
 Resources
Resources Protocol —— resDetect™ E. coli resDNA Quantitative Kit (qPCR)
Protocol —— resDetect™ E. coli resDNA Quantitative Kit (qPCR)
 Brochure —— resDetect™ CMC Process Residual Detection Solutions
Brochure —— resDetect™ CMC Process Residual Detection Solutions
 FAQ
FAQOPA-R005, the resDetect™ resDNA Sample Preparation Kit (Magnetic Beads), is recommended Its elution buffer is designed to optimize sample amplification efficiency and recovery rates in qPCR detection systems.
The standards for E. coli DNA are sourced from ECACC genomic DNA, and quantified using Qubit-4. Traceability is ensured through comparison with USP standard values.
High sensitivity is achieved through three main approaches: 1) optimization of trace nucleic acid extraction methods, 2) effective modification of primers and probes, 3) system optimization for optimal performance.
HEK293 cells, a versatile human cell line, are renowned for performing essential post-translational modifications like glycosylation and phosphorylation. These modification are crucial for human protein function, Making HEK293 cells invaluable in producing recombinant proteins such as monoclonal antibodies, cytokines, enzymes, and receptors.
The resDetect™ High-Sensitivity, High-Specificity HEK293 resDNA Quantitation Kit (qPCR), is designed for detection and quantification of residual host cell DNA from HEK293 cells! With high sensitivity, superior specificity, and optimization for quality control and regulatory compliance, our kit ensures the purity and safety of your recombinant proteins.
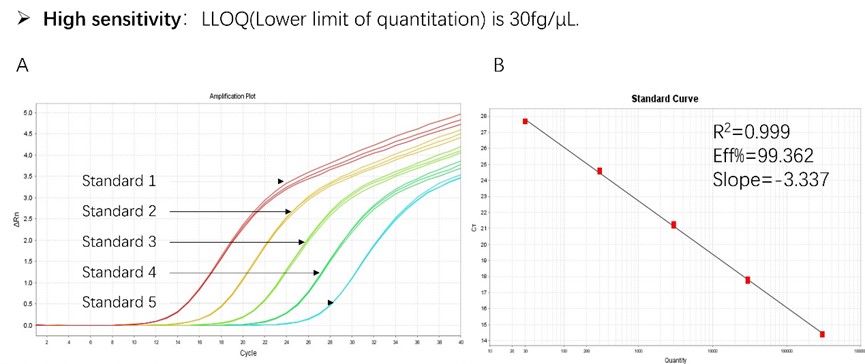
Figure 1. High sensitivity and broad dynamic range using the HEK293 resDNA Quantitation Kit. (A) Typical analysis results obtained with Standard 1 (300 pg/μL) to 5 (30 fg/μL). (B) The standard curve of the 10-fold dilution series. PCR efficiency should be 90-110%.
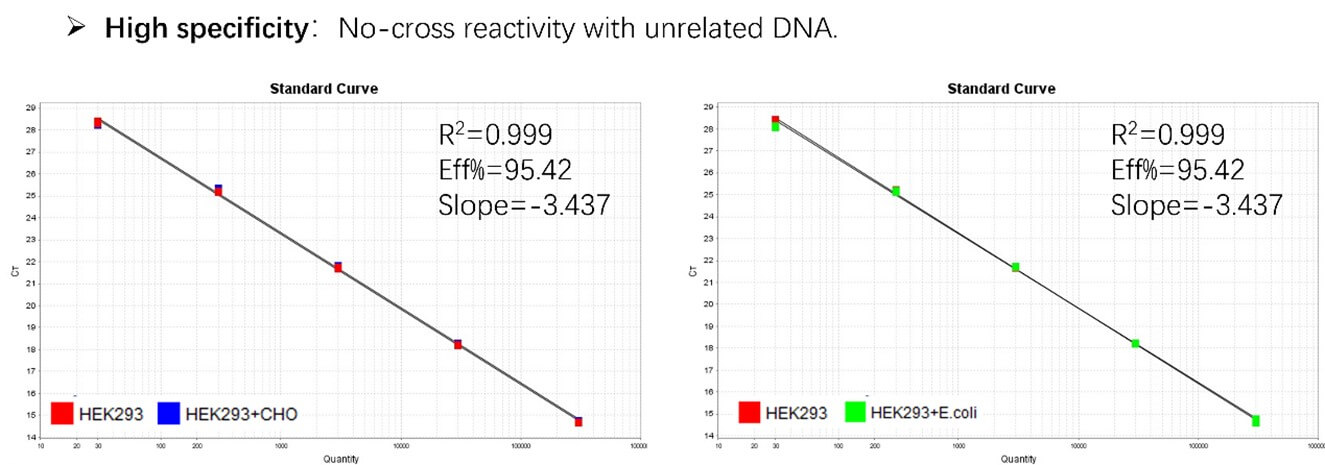
Figure 2. Assay specificity. Standard curves generated using 10-fold serial dilution of HEK293 genomic DNA (included in the kit) in the presence of CHO DNA and E. coli DNA.
 Products
Products resDetect™ HEK293 resDNA Quantitation Kit (qPCR)
resDetect™ HEK293 resDNA Quantitation Kit (qPCR)
 Resources
Resources Protocol —— HEK293 resDNA Quantitation Kit (qPCR)
Protocol —— HEK293 resDNA Quantitation Kit (qPCR)
 Brochure —— resDetect™ CMC Process Residual Detection Solutions
Brochure —— resDetect™ CMC Process Residual Detection Solutions
 FAQ
FAQWe recommend OPA-R024, resDetect™ resDNA Sample Preparation Kit II (Magnetic Beads). Its elution buffer is optimized to align with the qPCR detection system's buffer, ensuring optimal sample amplification efficiency, recovery rates, and differentiation of NTC.
Buffer NT and Buffer LA should not be premixed to avoid high salt precipitation, which could reduce extraction efficiency. However, Buffer NT can be safely premixed with Proteinase K to save time during experimental procedures.
Refer to the data analysis section of each kit's instruction manual for guidance. Threshold and baseline settings are determined based on the qPCR reaction system and the characteristics of different instruments. These settings are important for eliminating interference signals and optimizing qPCR amplification curves fitting.
Affinity chromatography is the standard for antibody drug purification, with Protein A widely used as a key ligand. Detecting residual Protein A is crucial to ensure the high-quality and safety of antibody drugs during process development and production. resDetect™ offers ready-to-use ELISA kits for detecting residues of natural Protein A, recombinant Protein A, MabSelect SuRe, Prism A, and Amsphere A3 fillers. We also provide customized kits for customers using Protein L and Protein G fillers, accommodating a wide range of antibody purification needs. Inquiry
Highly Recommended —— resDetect™ Universal Protein A Quick ELISA kit (Boiling-free)

![]() Universality: Detects both natural and structurally conserved recombinant Protein A, as well as common alkali-resistant Protein A mutants like MabSelect SuRe™ and Prism A.
Universality: Detects both natural and structurally conserved recombinant Protein A, as well as common alkali-resistant Protein A mutants like MabSelect SuRe™ and Prism A.
![]() Simple Sample Preparation: Requires no boiling or centrifugation , saving valuable time.
Simple Sample Preparation: Requires no boiling or centrifugation , saving valuable time.
![]() Method Validation: ICH-compliant validation reports available upon request.
Method Validation: ICH-compliant validation reports available upon request.
![]() High IgG Concentration Tolerance: Provides accurate quantification of antibodies up to 20 mg/mL.
High IgG Concentration Tolerance: Provides accurate quantification of antibodies up to 20 mg/mL.
The minimum detectable concentration of different forms of protein A are 40 pg/mL. The minimum detectable concentration was determined by adding twice standard deviations to the mean optical density value of twenty zero standard replicates and calculating the corresponding concentration.
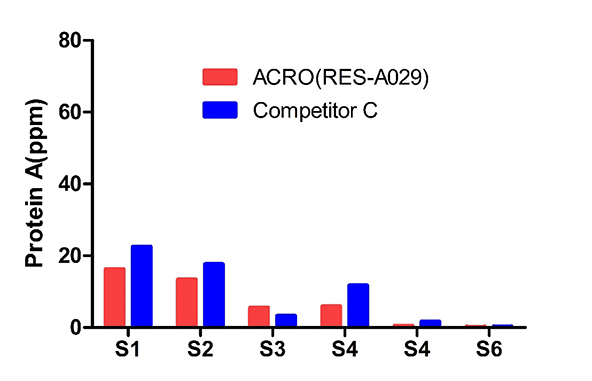
The sample batch samples (6) were tested by ACRO and Competitor C’s Kits. The Protein A final concentration ratio between them is 0.6-3.4.
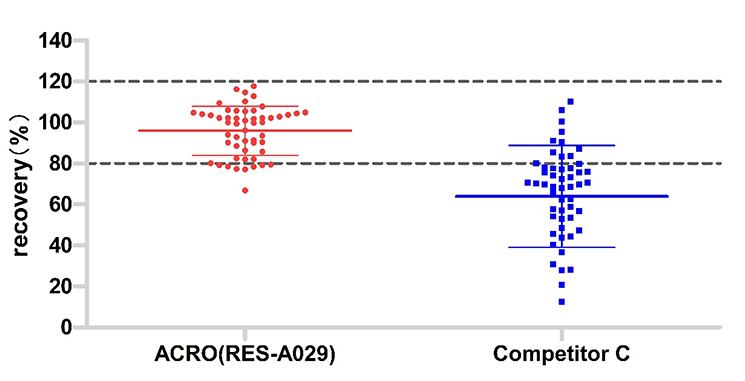
The same lot (53) samples were tested by those two kits. The acceptable recovery rate of ACRO (RES-A029) was 85%, and competitor C was 21%.
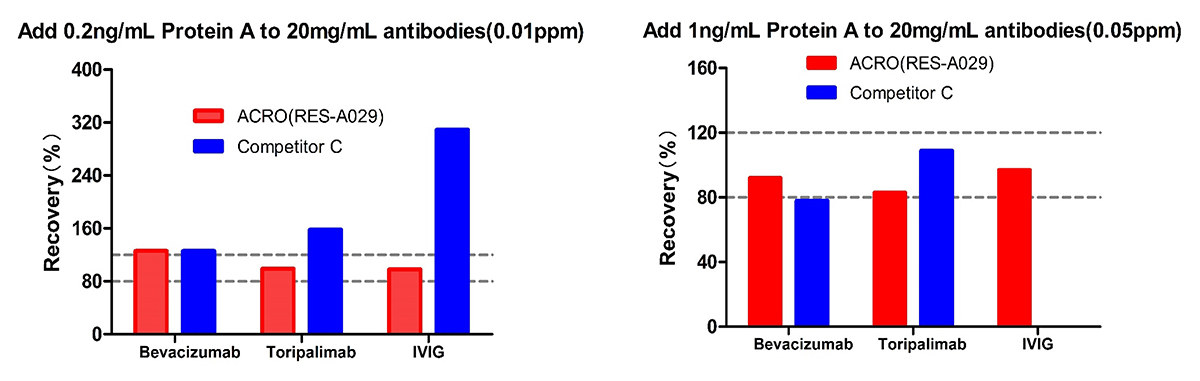
For Bevacizumab and Toripalimab, at low spiked concentration of Protein A (0.01ppm) level, Acro is better than Competitor C; meanwhile at high spiked concentration of Protein A (0.05ppm) level, the recovery rate of both are acceptable. For IVIG, the recovery rate for competitor C is not acceptable at both low and high spiked concentration of Protein A. But the recovery rate performance of ACRO are good at both levels. The results indicated that ACRO (RES-A029) was well tolerated when detecting Protein A residue in 20mg/mL IVIG.
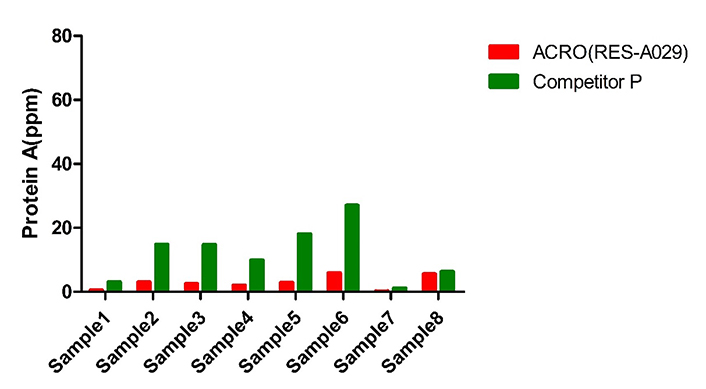
The same batch of sample (8) were tested by ACRO (RES-A929) and competitor P’s kit. The Protein A final concentration ratio between them is 1.1-10.9.
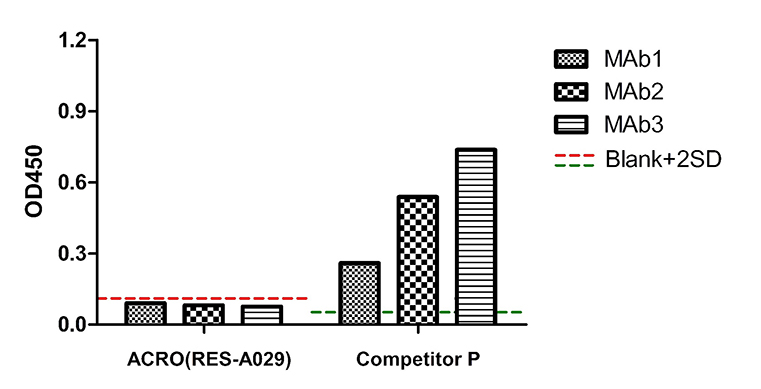
3 samples were purified by Protein G, and no Protein A residue since they were not purified by Protein A. The OD450 signal of 3 samples tested by ACRO(RES-A029) was below the blank+2SD. It means there is no Protein A residue in those samples. But the OD450 signal of the same samples tested by Competitor P was far beyond the blank+2SD. It means there are non-specific binding of Competitor P kit.
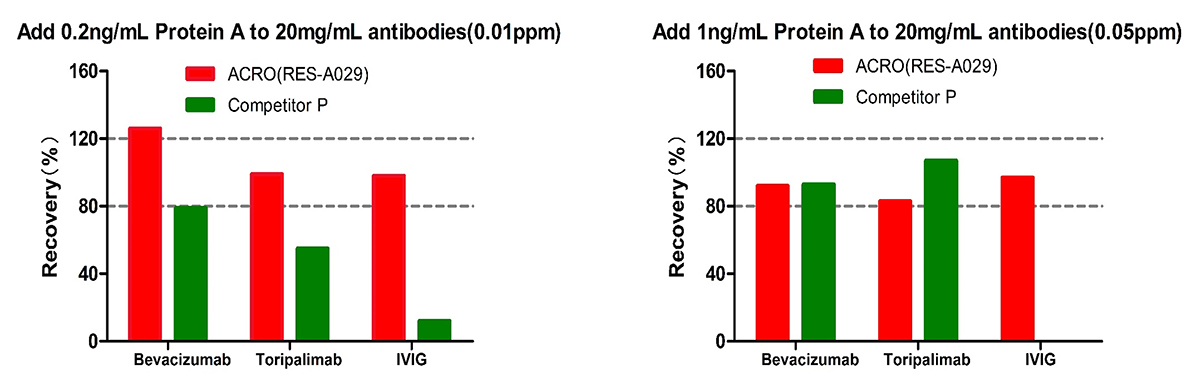
For Bevacizumab,the recovery rate for both kits are acceptable at low and high spiked Protein A level.
For Toripalimab, at low spiked concentration of Protein A (0.01ppm) level, Acro is better than Competitor P; meanwhile at high spiked concentration of Protein A (0.05ppm) level, the recovery rate of both are acceptable.
For IVIG, the recovery rate for competitor P is not acceptable at both low and high spiked concentration of Protein A. But the recovery rate performance of ACRO are good at both levels. The results indicated that ACRO (RES-A029) was well tolerated when detecting Protein A residue in 20mg/mL IVIG.
 FAQ
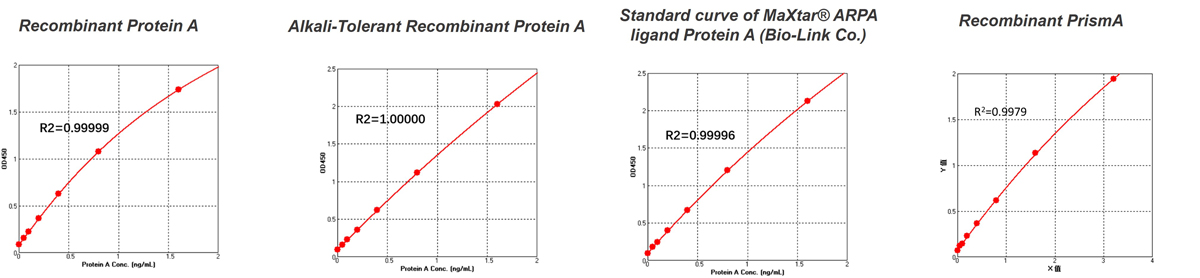
FAQThis assay kit is suitable for detecting natural or structurally conserved recombinant Protein A, including alkali-resistant recombinant Protein A used in bioprocess manufacturing ,such as MabSelect SuRe™, MaXtar® ARPA ligand (from Bio-Link). For other types of Protein A mutant variants used in affinity chromatography resins for antibody purification, it is recommended to obtain corresponding Protein A mutant standard protein solutions from the resin supplier. These standards should be stored under recommended conditions and diluted according to the assay kit instructions to fall within the concentration range of the standard curve for quantification. If the resin supplier cannot provide the specific Protein A mutant standard protein solutions, you may use the Protein A standard in the kit that is closest in characteristics for standard curve establishment and testing.
According to USP 1103 requirements for enzyme-linked immunosorbent assay (ELISA) methods: "Quantitative assays determine the quantity of the analyte based on the interpolation of a standard calibration curve with known analyte concentration, run simultaneously in the same assay. This standard should be an appropriate, preferably homologous, reference or calibration material that is representative of the analyte(s) of interest." Regulatory agencies recommend using matched standards to ensure the accuracy and reliability of assay results.
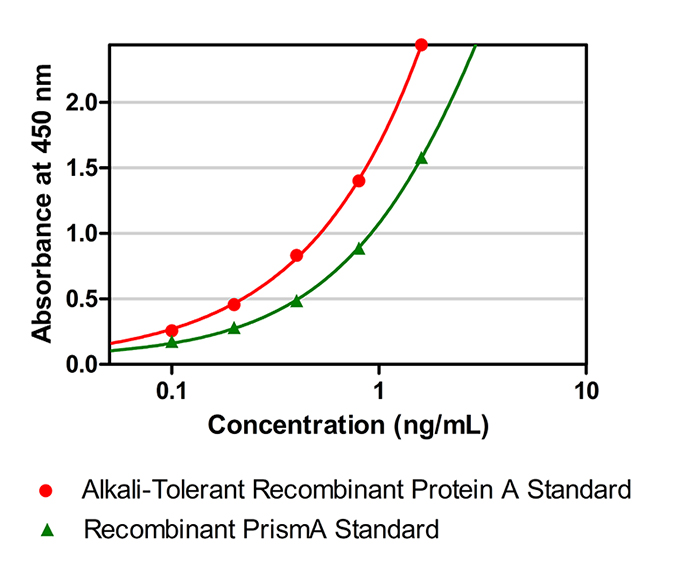
Figure 1. Standard curves plotted with standards from different sources
When comparing absorbance values plotted with Protein A from a generic kit and PrismA standards (see Figure 1), significant differences become evident. This discrepancy illustrates that accuracy in detecting residual ligands released from PrismAis substantially reduced when using generic standards for samples purified with PrismA. Therefore, to achieve more accurate results, it is recommended to use standards that match the purification fillers used in your process for calibrating sample measurements.
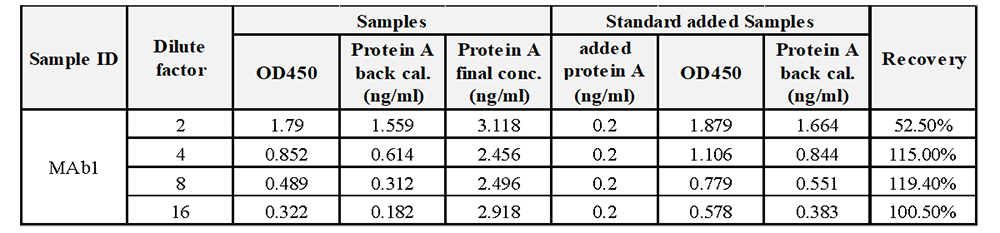
For samples containing Protein A residuals, where the residual amount cannot be predicted beforehand, it is recommended to start by diluting the samples at low dilution factors. At each dilution factor, spike the sample with a standard at a midpoint concentration to perform spiked sample detection and calculate the spike recovery rate. Ideally, the spike amount should closely match or equivalent to the analyte's concentration, with spike amount not exceeding three times the analyte's concentration. Post spiking, the measured value should not exceed 90% of the method's upper limit of quantification.
For example, if the Protein A residual value in a sample diluted 2-fold measures at 1.559, nearing the quantification limit, and when spiked with 0.2 ng/mL, the spike amount is notably lower than the residual amount, impacting accuracy in recovery rate. Through increasing the dilution factor, the measured value declines post dilution, nearing the spike amount and thereby ensuring acceptable spike recovery rates.
This web search service is supported by Google Inc.
