Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
The spike protein of SARS-CoV-2 facilitates viral entry into the target cell exploiting the Angiotensin-converting enzyme 2 (ACE2) receptor. The ACE2 gene is highly conserved in mammalian animals at the DNA and peptide level, which makes them the potential hosts for SARS-CoV-2.
A recent study found that the ACE2 gene was discovered in 12 common mammals. Among these 12 mammals, humans are the main host of SARS-CoV-2; mice are commonly used experimental animal models; bats and pangolins are suspected to be the intermediate host; cats, dogs, ferrets, and hamsters are pets; cattle, pigs, rabbits, and goats are common livestock. [1] Besides these 12 mammals, paguma larvata, an important host of SARS, also has ACE2 expression [2].
Although ACE2 is expressed in mice, they are not in any type of lung cells, but only in the tongue and skin. It is inferred that the expression pattern of ACE2 protein in mammals may be species-specific. There are amino acid residues on the binding surface of ACE2 which are essential for virus recognition. Mutations in these key amino acid residues may lead to the sensitivity change of cross-species infection. Seven critical differences were identified between the sequence of human ACE2 and mouse or rat ACE2. This could be the reason why mouse or rat ACE2 protein won’t bind to SARS-CoV-2 RBD protein. Thus, rodent models such as rats and mice may not be suitable for SARS-CoV-2 studies.
Studies have reported that the ACE2 gene is highly expressed on the skin and cornea of dogs. This could make dogs highly susceptible to SARS-CoV-2. Moreover, as you can see in the ACE2 phylogenetic tree below (Fig. 1), dogs are closer to humans than other mammals, indicating that they are more suitable as an animal model for COVID-19 study [1].
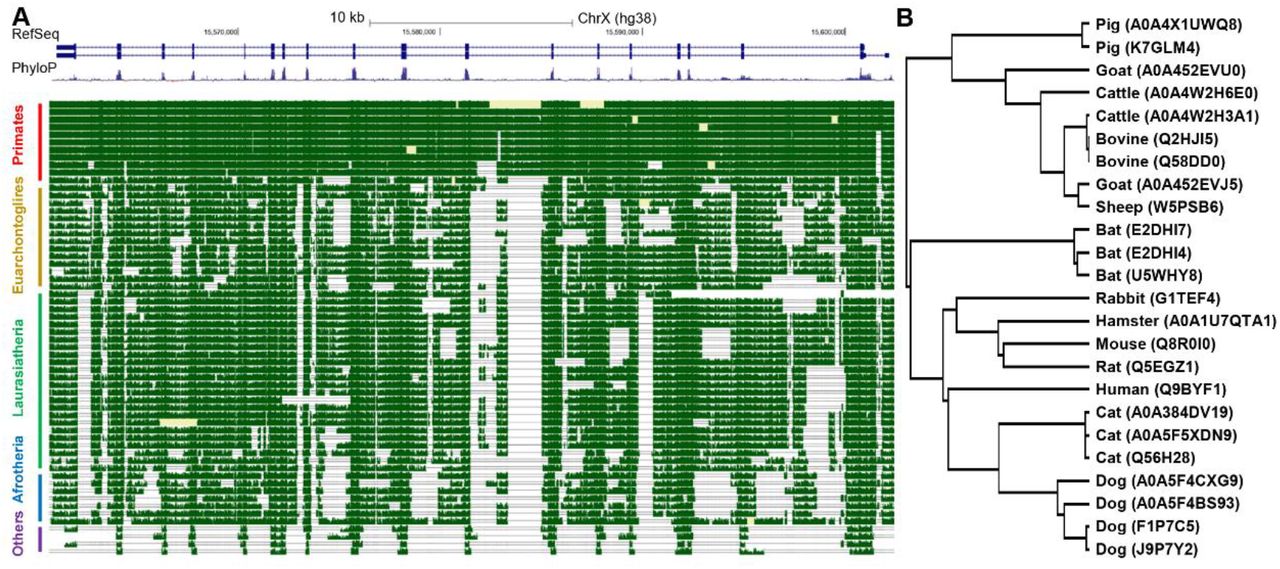
Fig. 1. Conservation analysis of ACE2 gene.
(Source: Sun et al., 2020 Atlas of ACE2 gene expression in mammals reveals novel insights in transmission of SARS-Cov-2, doi: https://doi.org/10.1101/2020.03.30.015644.)
Besides dogs, monkeys and apes are also susceptible to SARS-CoV-2. Several infection studies have shown that rhesus monkeys could be successfully infected with SARS-CoV-2, and subsequently develop COVID-19 like symptoms. The Cynomolgus ACE2 (Cat. # AC2-C52H7) developed by ACRO has been verified to have good binding activity with SARS-CoV-2 S1 protein and S RBD protein, which also confirms that cynomolgus ACE2 is highly homologous with Human ACE2 [3].
In summary, monkeys and dogs are better animal models for COVID-19 research.
Paguma larvata is an important host of SARS. Its ACE2 receptor protein also showed a very good binding ability to the S protein of SARS-CoV-2, though not comparable to human and cynomolgus ACE2. We did an ELISA assay to confirm this result. As you can see from the Fig. 2 below, the EC50s of paguma larvata ACE2 to SARS-CoV-2 S1 protein and S RBD protein were 52nM and 67nM, respectively. The EC50s of cynomolgus ACE2 to SARS-CoV-2 S1 protein and S RBD protein were 0.1 nM and 0.02 nM, respectively. The EC50s of human ACE2 to SARS-CoV-2 S1 protein and S RBD protein were 1.15 nM and 1.16 nM, respectively.
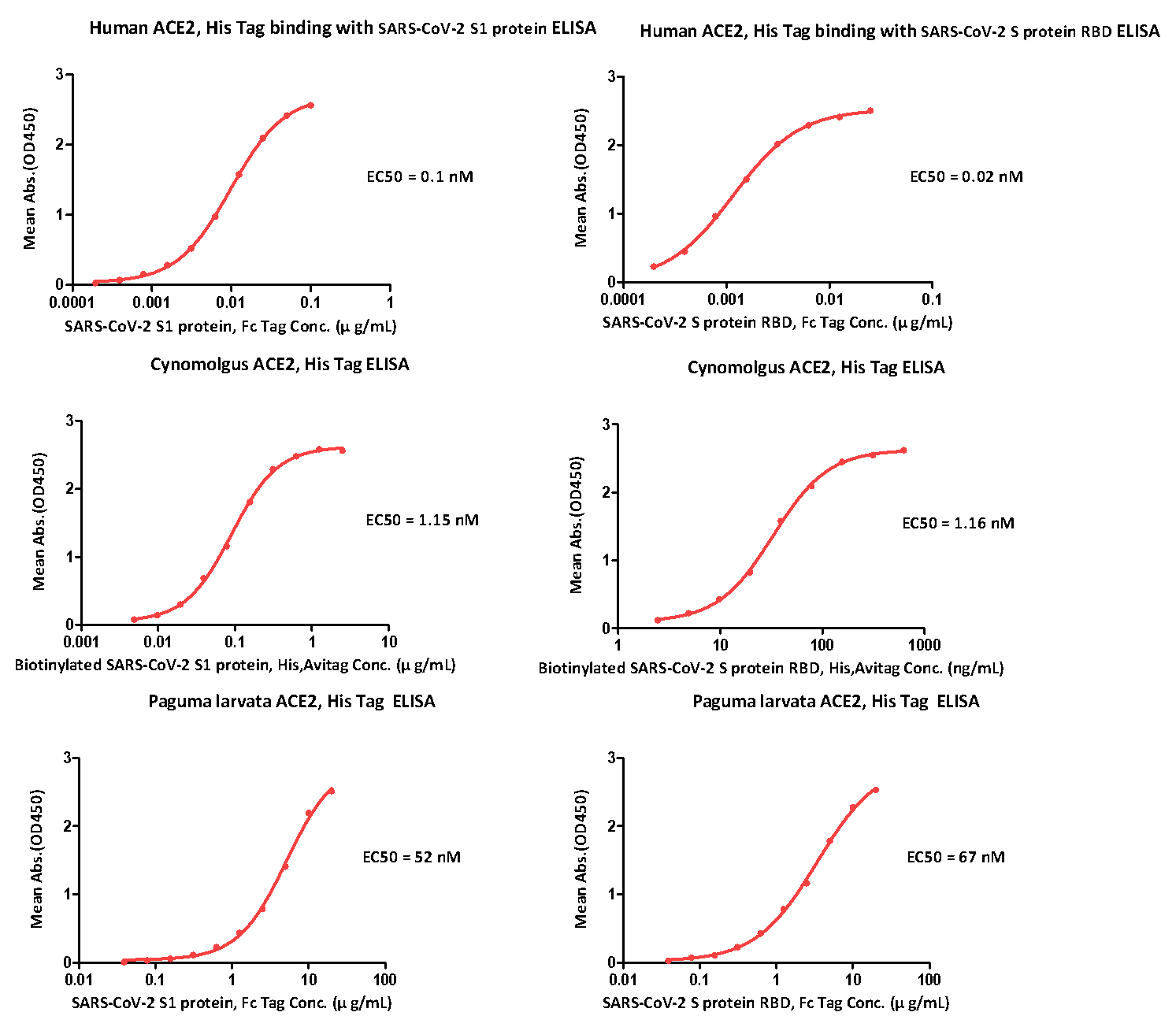
Fig.2 ELISA Binding activity results
We investigated the binding activity between the ACE2 of human, cynomolgus or paguma larvata, and the S RBD of SARS-CoV-2 using BLI. The results are summarized in Fig.3. The binding activities of human and cynomolgus ACE2 to the S RBD of SARS-CoV2 are much higher than paguma larvata ACE2.
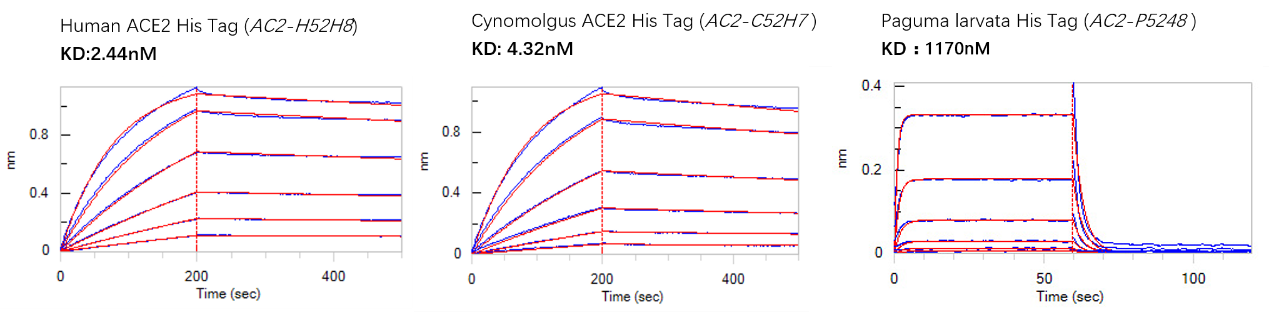
Fig.3 BLI binding activity results
An SPR method was also used to investigate the binding affinity between paguma larvata ACE2 and S RBD or S1 protein of SARS and SARS-CoV-2. Based on the results in Fig.4, we concluded that paguma larvata ACE2 is more susceptible to SARS.
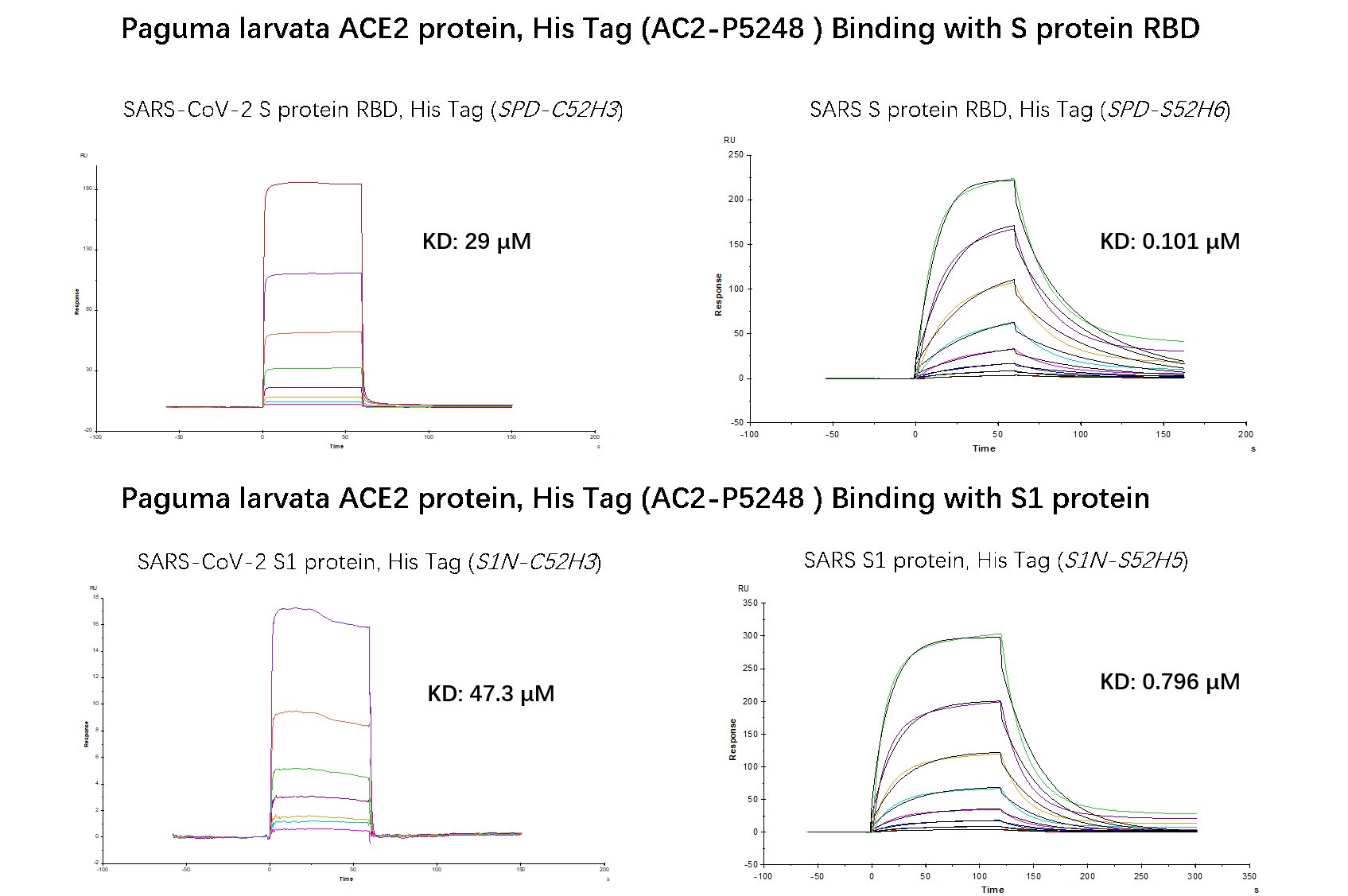
Fig.4 SPR binding activity results
ACROBiosystems has successfully developed ACE2 proteins for cynomolgus, rat, mouse, paguma larvata, human. The binding activity of these proteins has been verified by BLI and these proteins can be used for anti-SARS-CoV-2 research. (see Fig.2 ELISA Binding activity results above).
We also investigated the binding activity between the ACE2 of human, cynomolgus, paguma larvata and the S RBD of SARS-CoV-2 using BLI. The results are summarized in Fig.3. The binding activities of human and cynomolgus ACE2 are much higher than paguma larvata.
| Molecule | Cat. No. | Species | Host | Product Description | Order/Preorder |
|---|---|---|---|---|---|
| ACE2 | AC2-H82E6 | Human | HEK293 | Biotinylated Human ACE2 / ACEH Protein, His,Avitag™ (MALS verified) | |
| AC2-H82F9 | Human | HEK293 | Biotinylated Human ACE2 / ACEH Protein, Fc,Avitag™ | ||
| AC2-H5257 | Human | HEK293 | Human ACE2 / ACEH Protein, Fc Tag (MALS verified) | ||
| AC2-C52H7 | Cynomolgus | HEK293 | Cynomolgus ACE2 / ACEH Protein, His Tag | ||
| AC2-H52H8 | Human | HEK293 | Human ACE2 / ACEH Protein, His Tag (MALS verified) | ||
| AC2-R5246 | Rat | HEK293 | Rat ACE2 / ACEH Protein, His Tag (MALS verified) | ||
| AC2-M5248 | Mouse | HEK293 | Mouse ACE2 / ACEH Protein, His Tag (MALS verified) | ||
| AC2-P5248 | Paguma larvata | HEK293 | Paguma larvata ACE2 / ACEH Protein, His Tag |
>>>More SARS-CoV-2 products and kits
Reference:
[1] Sun et al., 2020 Atlas of ACE2 gene expression in mammals reveals novel insights in transmisson of SARS-Cov-2, doi: https://doi.org/10.1101/2020.03.30.015644
[2] Stawiski et al., 2020 Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. doi: https://doi.org/10.1101/2020.04.07.024752
3] Melin et al., 2020 Comparative ACE2 variation and primate COVID-19 risk, https://doi.org/10.1101/2020.04.09.034967
4) Wrapp et al., 2020 Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. DOI: 10.1126/science. Abb 2507
This web search service is supported by Google Inc.







