Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> Insights > S protein is in a trimeric form as verified by three methods from NIH SARS-CoV-2 is a positive-sense single-stranded RNA virus that is very contagious in humans. The viral envelope consists of a lipid bilayer, in which the membrane (M), envelope (E) and spike (S) structural proteins are anchored. The spike protein is a homotrimer, which is composed of an S1 and S2 subunit. The globular S1 subunit is involved in receptor recognition, whereas the S2 subunit facilitates membrane fusion and anchors S into the viral membrane. Therefore, the S protein is the key protein that the coronavirus uses to invade human cells. S protein is also the main antigen that causes the host immune system to produce neutralizing antibodies after infection.
According to the article published in Science earlier this year, a team from UT Austin determined the cryo-electron microscopy structure of the 2019-nCoV S trimer in the prefusion conformation. [1] As shown in Fig.1, prefusion S glycoproteins adopt a similar mushroom-like homo-trimer architecture, of which the stem is mainly composed of three S2 subunits, and the top cap consists of three interwoven S1 subunits.[3] Another team from Tsinghua University confirmed that the conformational switch of the receptor-binding domain(RBD) from the “down” to “up” position is a prerequisite for receptor binding (Fig. 2). Therefore, the homo-trimer architecture is essential to achieve the complete function of the S protein.
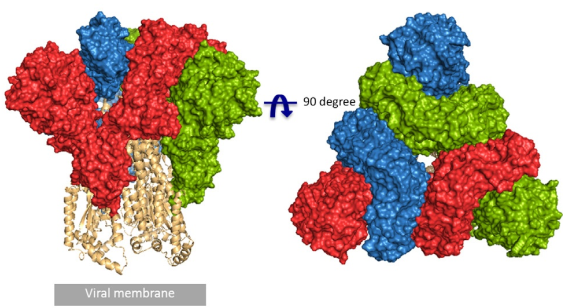
Fig. 1 Front and top view of the trimeric coronavirus spike protein ectodomain obtained by cryo-electron microscopy analysis. Three S1 protomers (surface presentation) are colored in red, blue, and green. The S2 trimer (cartoon presentation) is colored in light orange. (Source: R.J.G. Hulswit, et al., Coronavirus Spike Protein and Tropism Changes. Adv Virus Res. 2016; 96: 29–57.)
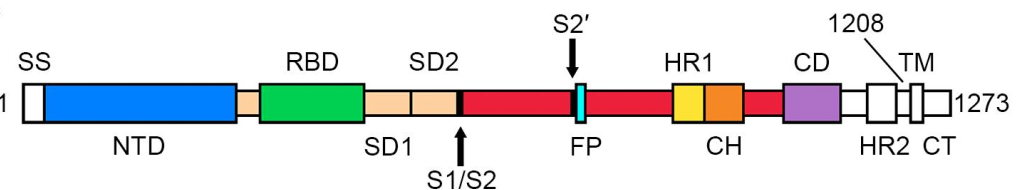
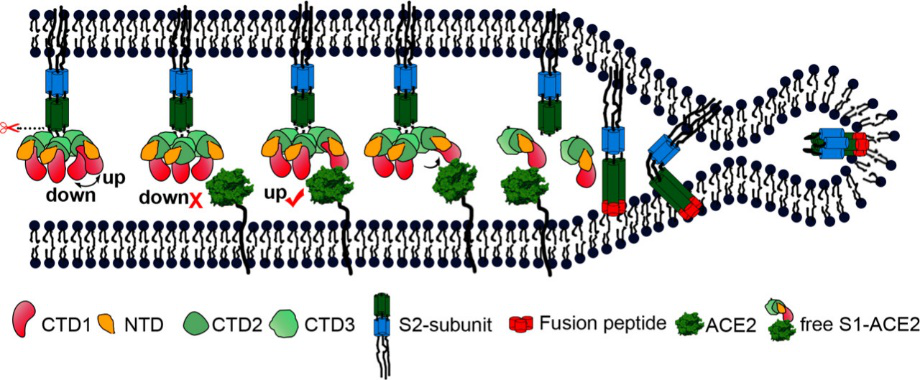
Fig. 2 S protein mediates viral membrane and membrane fusion by binding to host cell receptor ACE2. (Source: Song W, Gui M, Wang X, Xiang Y (2018) Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 14(8): e1007236.)
The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics. The correct trimeric structure and functionality of the recombinant S protein reagent are important. Therefore, strict quality control is required in terms of the molecular weight and aggregation state. In a recent work from NIH, the scientist used three ways including SDS-PAGE, analytical size exclusion chromatography, and transmission electron microscopy to verify that the recombinant S protein is in a trimeric form when developing a COVID-19 serological diagnostic kit (Fig. 3).
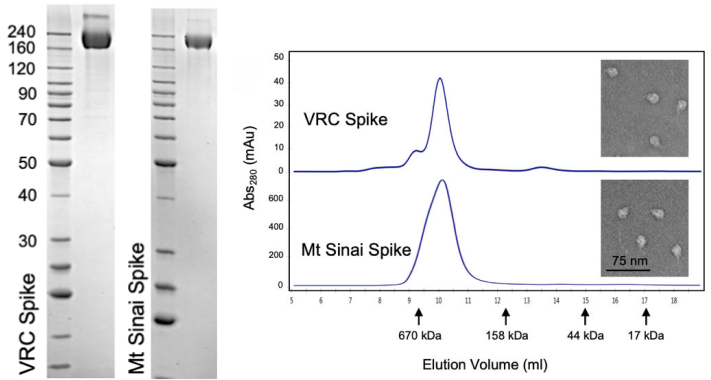
Fig. 3 Verification of Spike trimer structure using SDS-PAGE, analytical size exclusion chromatography and transmission electron microscopy
(Source: Carleen Klumpp-Thomas, et al. Standardization of enzyme-linked immunosorbent assays for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling, preprint at https://www.medrxiv.org/content/10.1101/2020.05.21.20109280v1.)
ACROBiosystems has specially designed a highly active S trimer protein. This is the only verified trimeric S protein on the market. Our SEC-MALS and negative-stain EM data reveal that ACRO's S protein is in the correct trimer form under physiological conditions, and purity is over 80%. The Biacore affinity test shows that the S trimer protein has an affinity constant as 35.3nM to human ACE-2 protein, which is similar to the value in the reference.[1]
Reference:
1. Wrapp D, et al., Cryo-EM structure of the 2019-nCoV spike prefusion conformation. Science, 2020 Mar 13;367(6483):1260-1263.
2. R.J.G. Hulswit, et al., Coronavirus Spike Protein and Tropism Changes. Adv Virus Res. 2016; 96: 29–57.
3. Song W, Gui M, Wang X, Xiang Y (2018) Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 14(8): e1007236.
4. Carleen Klumpp-Thomas, et al. Standardization of enzyme-linked immunosorbent assays for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling, preprint at https://www.medrxiv.org/content/10.1101/2020.05.21.20109280v1.
This web search service is supported by Google Inc.







