 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> Insights > Can aerosolized nanobodies be a gamechanger in the fight against COVID-19? After the outbreak of SARS-CoV-2, researchers as well as biotech and pharma industry responded quickly to develop vaccines and therapeutics. Within less than a years’ time, breakthroughs in research and developments brought multiple vaccines and antibody therapeutics to hospitals at an unprecedented rate.
Among the most successful COVID-19 therapies are neutralizing antibodies that block viral entry into host cells and reduce severity and length of the disease. The FDA has given 2 emergency use authorizations for monoclonal antibodies developed by Regeneron and Eli Lilly. More than 10 new antibodies are currently in clinical trials. These immunoglobulin (IgG) antibodies contain variable heavy (VH) and variable light (VL) antigen binding domains which makes them large and increases amounts needed for therapy use in addition to limiting delivery to intravenous (IV) drips. However, some mammals including alpacas, camels and related species carry distinct antibodies containing only a VHH domain but lacking VL domain. The VHH domains by itself, termed nanobodies, have high water solubility, stability, improved tissue penetration and are amenable to genetic engineering. Furthermore, nanobodies are one tenth the size of an IgG antibody which lowers manufacturing cost and can be delivered to patients via aerosolized particle in nasal spray.
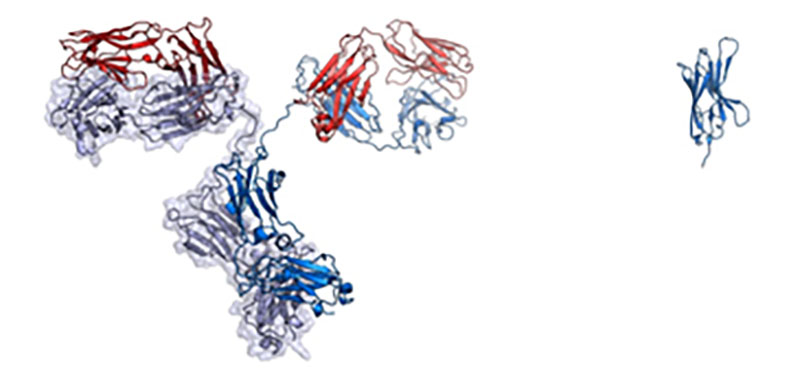
On the left is a full-sized IgG antibody while on the right is a nanobody.
Credit: Walter and Manglik Labs/UCSF/HHMI
On December 22, 2020, scientists at the NIH published on a nanobody, NIH-CoVnb-112, derived from a llama named Cormac that can effectively neutralize SARS-CoV-2. Furthermore, the researchers explored impact of aerosolization on the nanobody activity.
The development process and evaluation results of this antibody are described as follows:
1. The researchers immunized an adult Llama with the commercially available SARS-CoV-2 S1 protein (Cat.S1N-C5255) and then isolated the B-cells from the llama blood and amplified single heavy-chain variable domain to build a 108 clones phage display library (Figure 1A). The researchers then screened nanobody clones using S protein RBD and ACE-2 competition method (Figure 1B): i) the human ACE2 (Cat. AC2-H52H8) was fixed to the radioimmunoassay tube, then a premix of nanobody clones incubated with biotinylated RBD (Cat. SPD-C82E9)was added to the tube; ii) nanobody that bound to RBD and blocked RBD/ACE-2 interaction were fished out of the solution via streptavidin coated beads; iii) the unbound nanobody to the streptavidin beads were washed and neutralizing nanobody bound to RBD were enriched and eluted.
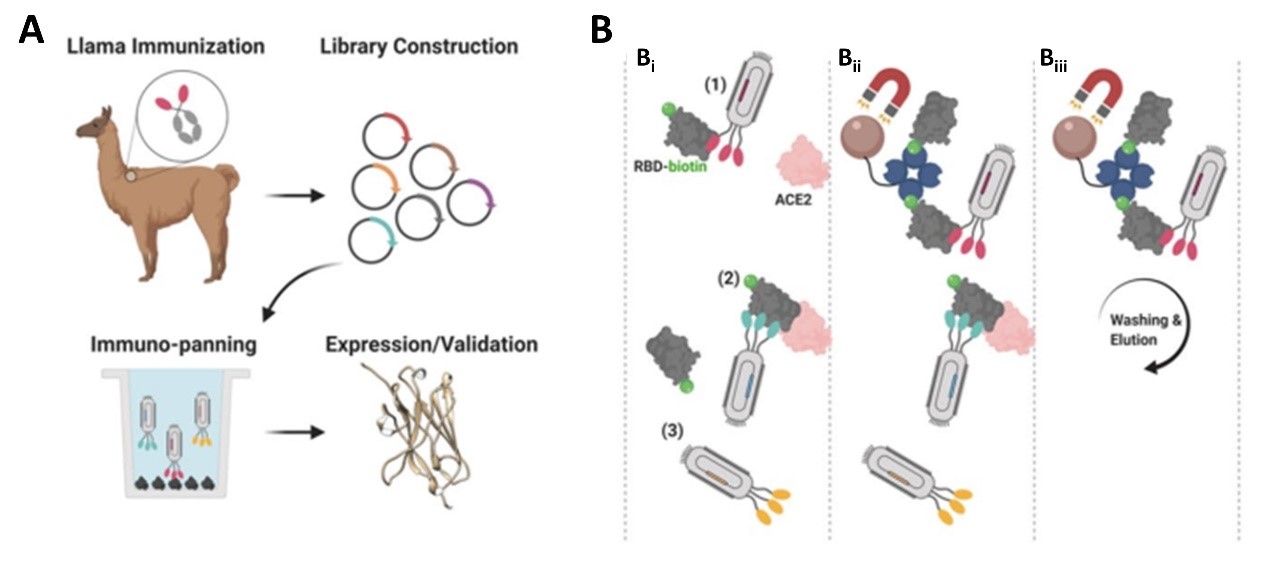
Figure 1. Nanobody development process. A) Llama immunization and identification of nanobody candidates. B) Selection of neutralizing nanobodies using three step process.
2. The researchers sequenced hundreds of nanobody clones and isolated 13 unique clones numbering from NIH-CoVnb-101 to NIH-CoVnb-113. These nanobodies were overexpressed in E. coli, purified and the affinity between nanobody and RBD (Cat. SPD-C82E9) was measured via biolayer interferometry (BLI). 5 out 13 nanobody displayed the highest affinity in the nanomolar range (Figure 2). The NIH-CoVnb-112 nanobody displayed the highest affinity to RBD with a KD = 4.9 nM (Figure 2).
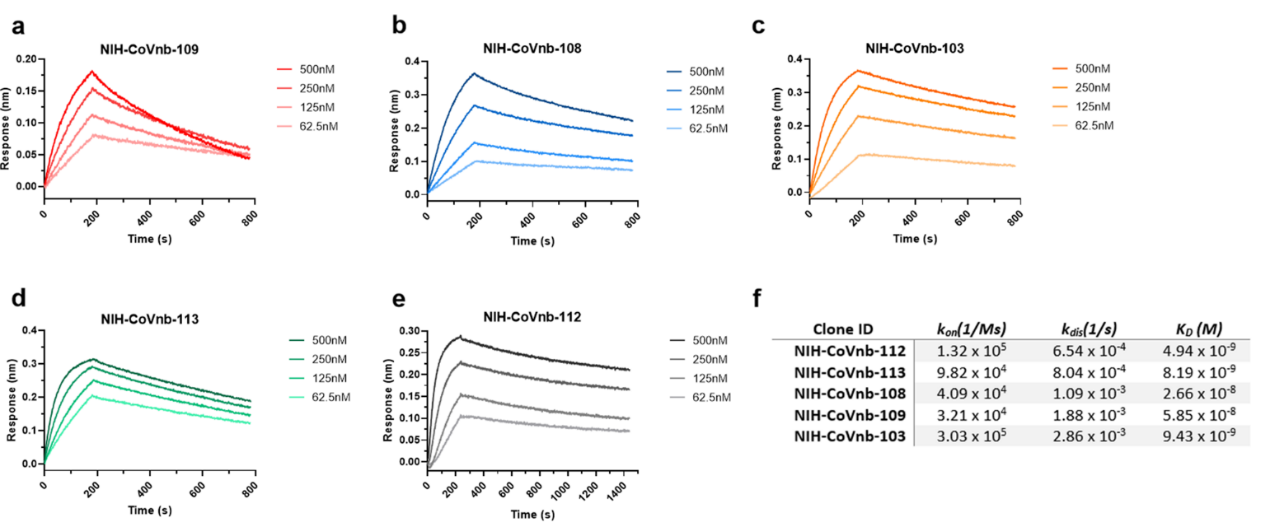
Figure 2. BLI analysis of RBD binding affinity of isolated nanobody clones.
3. In-vitro competitive experiments have shown that screened nanobodies block RBD/ACE2 binding with NIH-CoVnb-112 having the lowest EC50 value at 1.11 nM (Figure 3)
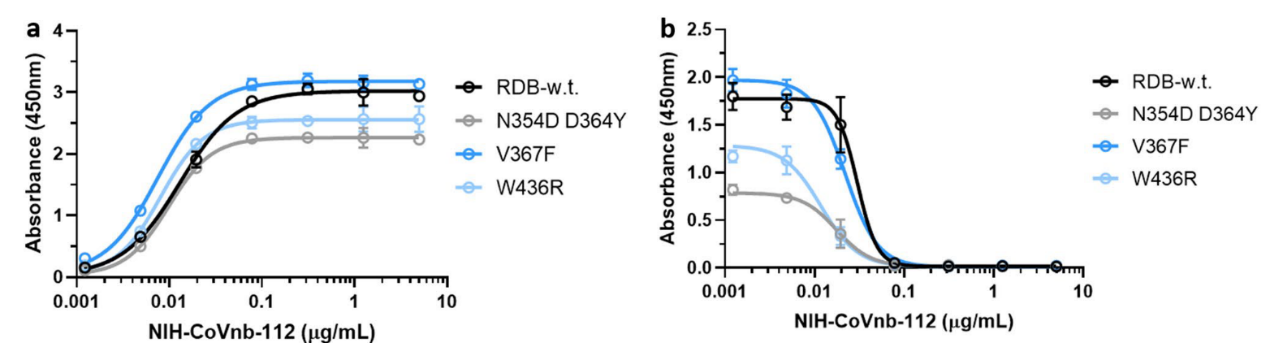
Figure 3. NIH-CoVnb-112 effectively suppresses ACE2/RBD binding
4. Mutations in the new coronavirus RBD region may help the virus escape vaccine induced immunity or lower efficacy of existing neutralization antibodies. Therefore, the researchers tested the interaction of NIH-CoVnb-112 with three RBD mutants (N354D/D364Y, V367F, W436R). No significant difference in binding affinity (Figure 4a) was observed compared to wild type RBD (Cat. SPD-C52H3). While competition inhibition assay using RBD coated plates incubated with NIH-CoVnb-112 blocked ACE2 interaction showed similar EC50 to each of the RBD mutants (Figure 4b). This suggests that NIH-CoVnb-112 may be able to effectively suppress infection with new SARS-CoV-2 containing these RBD mutations.
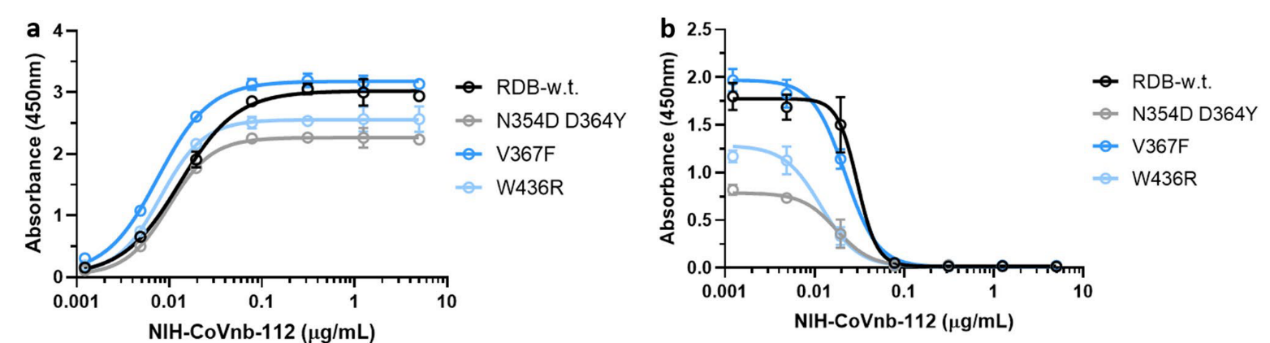
Figure 4. NIH-CoVnb-112 binds to RBD mutants and effectively inhibits ACE2/RBD mutant binding
5. The binding affinity of NIH-CoVnb-112 to SARS-CoV-1 RBD was also assessed using BLI assay (Figure 5a) and ELISA (Figure 5b). Both assays determined that NIH-CoVnb-112 did not bind to SARS-CoV-1 RBD.
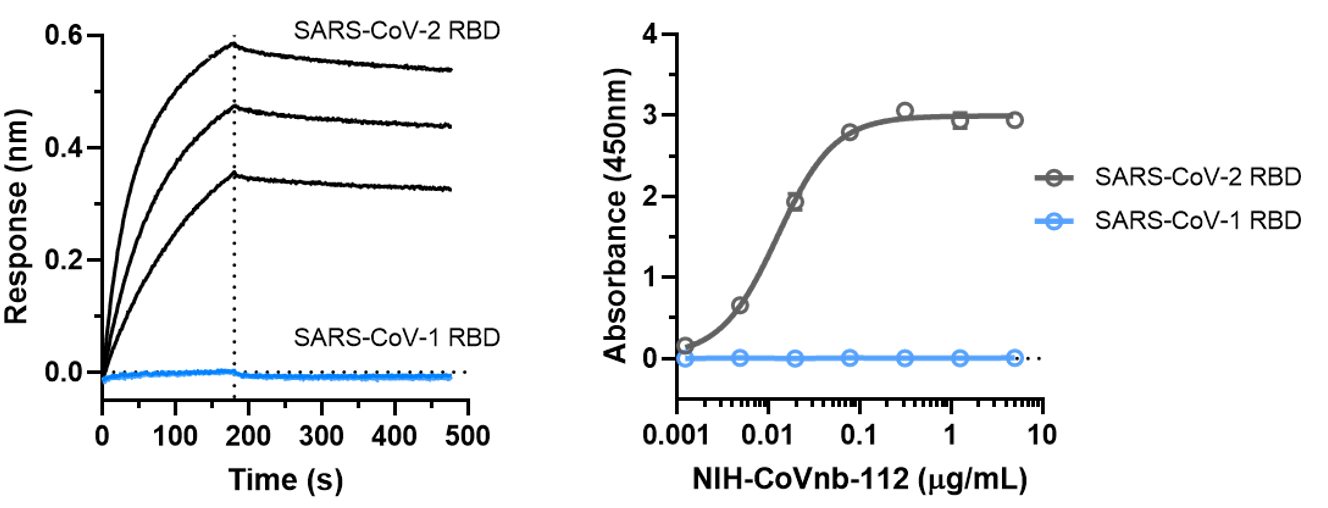
Figure 5. NIH-CoVnb-112 does not bind to SARS-CoV-1
6. A study of the stability of NIH-CoVnb-112 by Circular Dichroism (CD) spectroscopy found that the structure of the nanobody began to unfold at 74.4 oC, and 73% of the nanobody refolded after temperature was lowered indicating high stability of the nanobody.
7. As a result of high stability and low molecular weight, nanobodies can be aerosolized. The authors tried to use commercial nebulizer to aerosolize NIH-CoVnb-112 (Figure 6a). SDS-PAGE and size exclusion chromatography analysis showed no degradation or aggregation of the nanobody after aerosolization (Figure 6b-C). To assess functional effect on the nanobody post nebulization, a fluorescence reporter assay using S protein pseudotyped lentiviral infection was used (Figure 6d). NIH-CoVnb-112 was able to inhibit viral transduction before and after aerosolization (Figure 6e). The combined results show that NIH-CoVnb-112 structure and activity is not affected by aerosolization process.
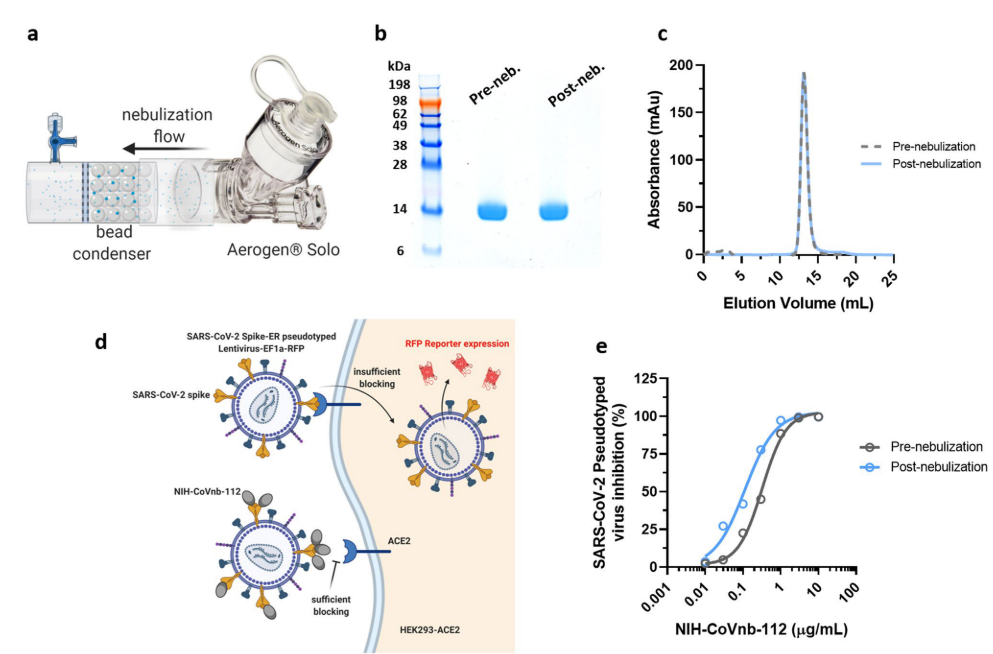
Figure 6. Aerosolized NIH-CoVnb-112 is structurally stable and can effectively block pseudotyped SARS-CoV-2 viral transduction.
The lead nanobody identified in this study, NIH-CoVnb-112, has the potential to be developed into a therapeutic against COVID-19. Further pre-clinical characterization is needed before bringing NIH-CoVnb-112 into clinical trial. However, ACROBiosystems is excited to have provided key protein reagents at each step of the work including S1 protein for immunization of the llama, biotin-RBD protein for nanobody clone screening, and ACE2 protein used in RBD/ACE2 interaction assays.
Before conducting the experiments, researchers verified the purity and activity of ACROBiosystem's SARS-CoV-2 products with satisfactory results (Figure 7a-c).
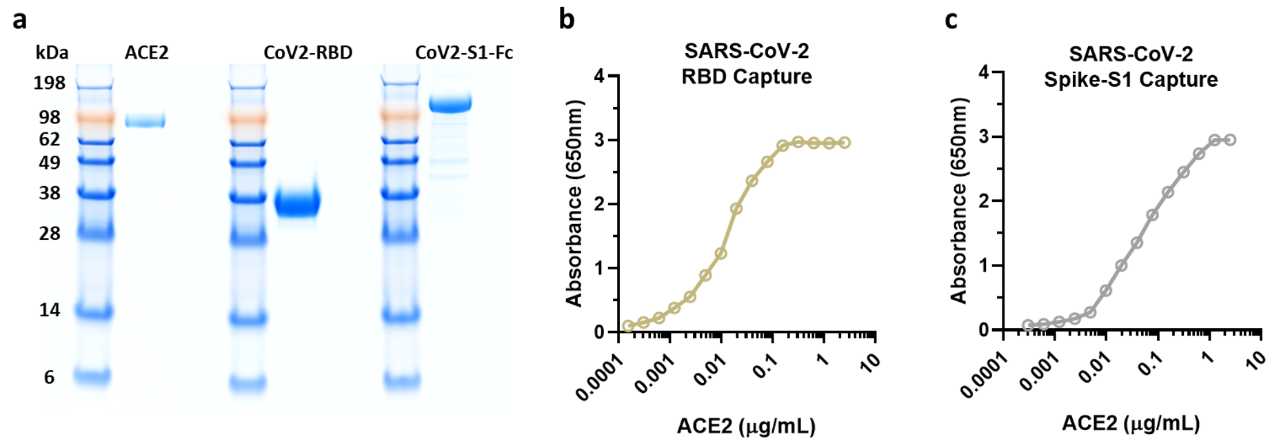
Figure 7. NIH researchers test ACRO protein purity and activity results
ACROBiosystems will continue to support this work as well as wider research and development work in the fight against COVID-19 with high quality protein and reagent products.
ACROBiosystems is committed to helping advance targeted therapeutic drugs, diagnostics, and vaccine development by providing high quality target antigens, key reagents and related service.
References:
1. Esparza TJ, Martin NP, Anderson GP, Goldman ER, Brody DL. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci Rep. 2020Dec 22; 10(1):22370. doi: 10.1038/s41598-020-79036-0. PMID: 33353972; PMCID: PMC7755911
2. Starr T N, Greaney A J, Hilton S K, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding[J]. Cell, 2020, 182(5): 1295-1310. e20.
3. Mulligan M J, Lyke K E, Kitchin N, et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults[J]. Nature, 2020: 1-5.
4. Rudberg A S, Havervall S, Månberg A, et al. SARS-CoV-2 exposure, symptoms and seroprevalence in healthcare workers in Sweden[J]. Nature communications, 2020, 11(1): 1-8.
5. Kondo T, Iwatani Y, Matsuoka K, et al. Antibody-like proteins that capture and neutralize SARS-CoV-2[J]. Science advances, 2020, 6(42): eabd3916.
6. Ravichandran S, Coyle E M, Klenow L, et al. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits[J]. Science Translational Medicine, 2020.
7. Hao W, Ma B, Li Z, et al. Binding of the SARS-CoV-2 Spike Protein to Glycans. BioRxiv preprint. doi: https://www.biorxiv.org/content/10.1101/2020.05.17.100537v1
8. Pierce C A, Preston-Hurlburt P, Dai Y, et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients[J]. Science translational medicine, 2020, 12(564).
9. Luetkens T, Metcalf R, Planelles V, et al. Successful transfer of anti–SARS-CoV-2 immunity using convalescent plasma in an MM patient with hypogammaglobulinemia and COVID-19[J]. Blood advances,2020, 4(19): 4864-4868.
10. Ropa J, Cooper S, Capitano M L, et al. Human hematopoietic stem, progenitor, and immune cells respond ex vivo to SARS-CoV-2 spike protein[J]. Stem cell reviews and reports, 2020: 1-13.
11. Lou Y, Zhao W, Wei H, et al. Cross-neutralization antibodies against SARS-CoV-2 and RBD mutations from convalescent patient antibody libraries. BioRxiv preprint. doi: https://doi.org/10.1101/2020.06.06.137513
This web search service is supported by Google Inc.
