Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
The SARS-CoV-2 virus consists of four structural proteins: spike protein (S), nucleocapsid protein (N), envelop protein (E), and membrane protein (M). Among them, S protein is responsible for recognizing the target receptor and mediating the fusion of the virus and the target cell membrane, which is considered as the key to the infection process. S protein is also the main target for neutralizing antibodies after infection. Therefore, compared to the other three structural proteins, S protein has attracted much more attention in the development of COVID-19 vaccines and therapeutic antibodies.
>>>Click to find out how SARS-CoV-2 infects the host
S protein contains two subunits, S1 and S2. During the infection process, the S1 subunit binds to the ACE2 receptor while the S2 subunit anchors the S protein to the host cell membrane and mediates the fusion of the virus envelope and the host cell membrane. There are two important domains in S1 subunits, known as N-Terminal Domain (NTD) and Receptor Binding Domain (RBD). We know that RBD is involved in binding to ACE2. S2 has three domains called Heptad Repeat (HR), Central Helix (CH), and Connector Domain (CD) respectively. Additionally, there is a furin cleavage site at S1/S2 (Fig. 1)1.
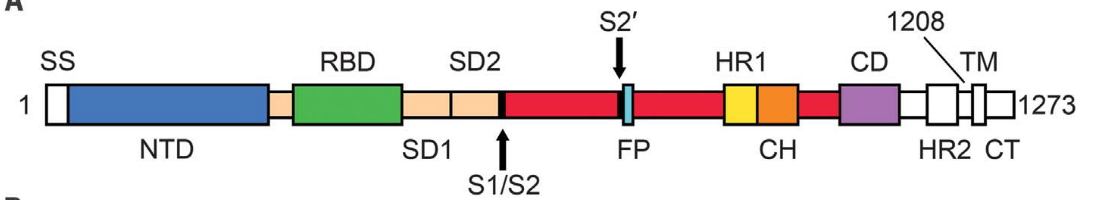
Fig. 1 Sequence of Spike protein
The S protein protrudes from the viral surface as a homotrimer with two different conformations, prefusion and postfusion (Fig. 2).2 The binding of RBD and ACE2 triggers the structural change, which dissociates the S1 and S2 subunits and transforms the S2 subunit into a highly stable post-fusion conformation. Scientists found that the S protein is unstable structurally. It will spontaneously change from prefusion to postfusion conformation even without binding, which poses a huge challenge to the development of vaccines and therapeutic antibodies.
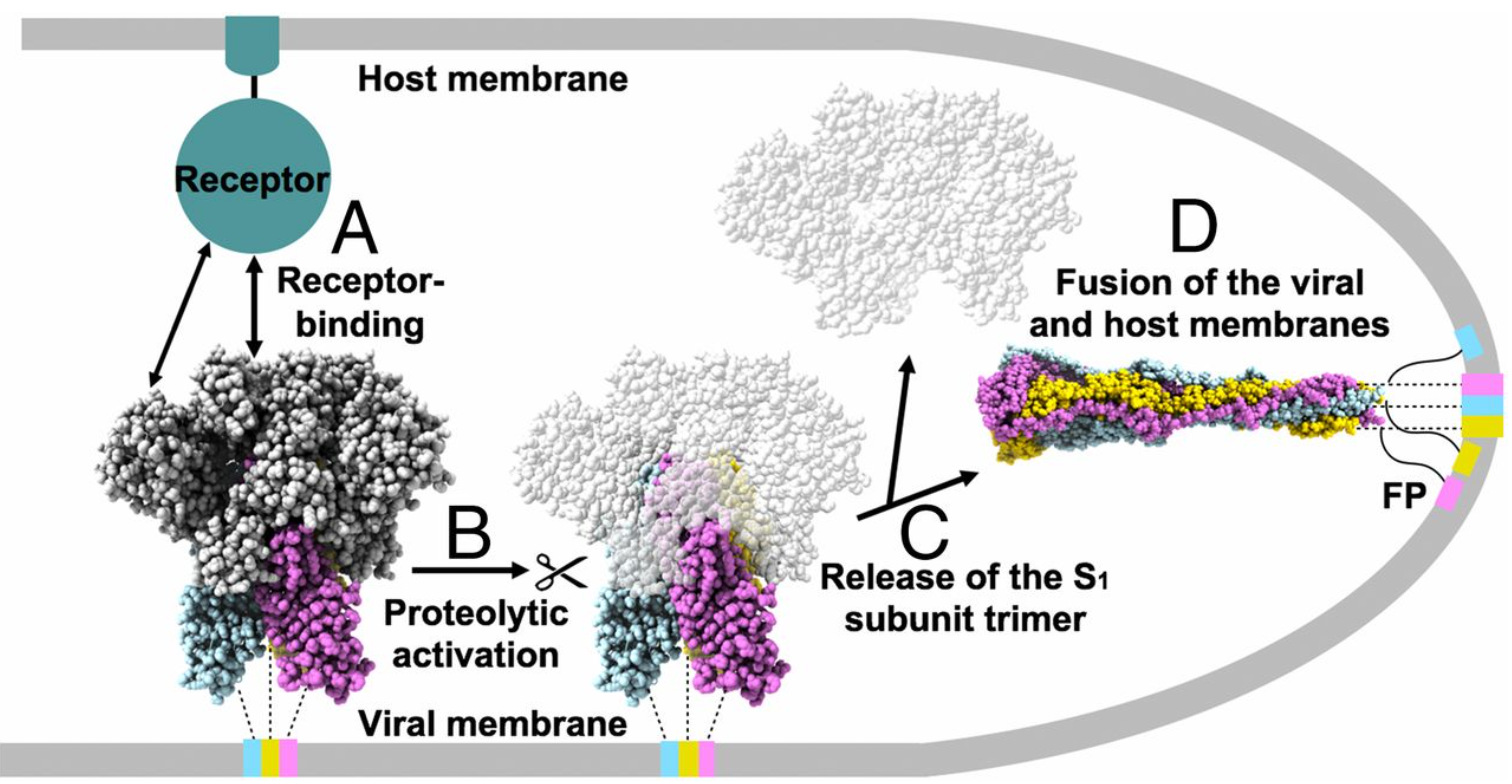
Fig. 2 Proposed model of coronavirus entry.
Considering the importance of S protein, it is going to be the key to get the stable and uniform trimeric S protein reagent for the development of vaccines, therapeutic antibodies, or serological test kits. After abundant work, scientists found that 2 proline mutations (S-2P) of the S2 subunit stabilize the S protein conformation, most of which will stay at prefusion state. We will walk you through a few different scenarios that recombinant spike trimer protein could help in the following part.
Scenario 1: Structural biology research
It is important to study the three-dimensional structure of the S protein, especially for the understanding of receptor recognition and membrane fusion process. The unstable conformation of natural S protein causes a lot of trouble. S-2P mutations stabilize the protein. Scientists can use recombinant S-2P protein samples to study the structural biology (Fig. 3).1
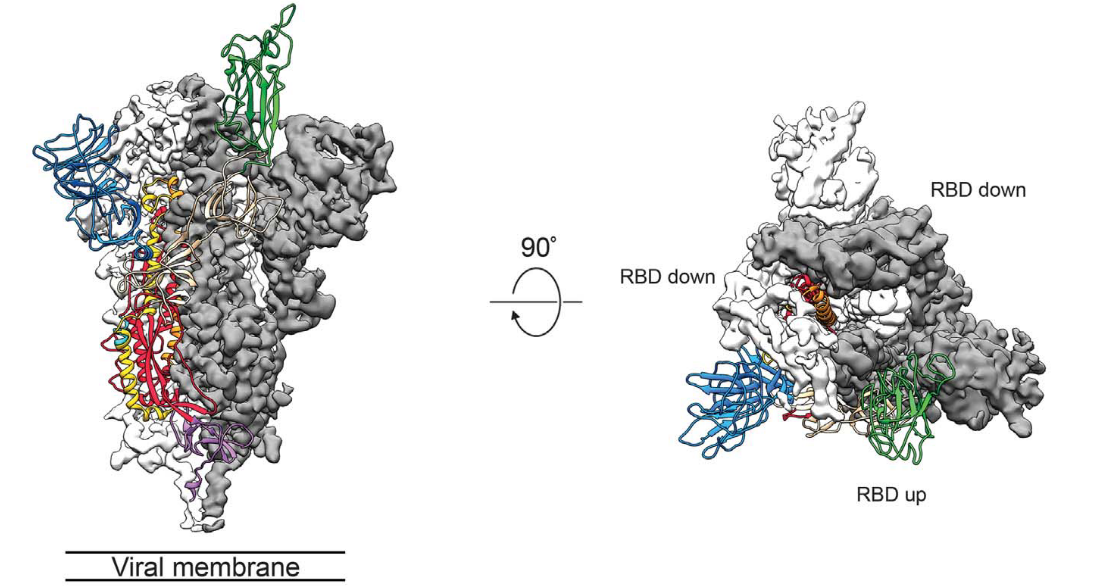
Fig. 3 Cryo-EM of Spike protein
>>>Click to find more SARS-CoV-2 related products
Scenario 2: Vaccine development
A vaccine typically contains an agent that resembles a disease-causing microorganism. The agent stimulates the body's immune system to produce neutralizing antibodies to protect from the agent that it may encounter in the future. Currently, there are more than 300 COVID-19 vaccines in different development stages worldwide. Except for the inactivated vaccines, most vaccines use S protein as the immunogen. The vaccine developed using S trimer protein in the prefusion state could better protect human from SARS-Cov-2 attack, since the virus is in the prefusion state when they attach to the target cell. We found that a lot of companies use S-2P in their vaccine design as well, including Moderna, Pfizer, Johnson&Johnson, and Novavax. This finding reflects the importance of the S protein conformation in the vaccine development(Fig. 4).
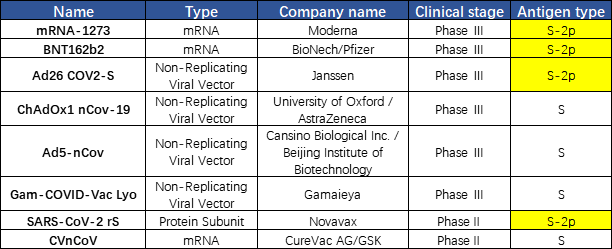
Fig. 4 Vaccines under development targeting S protein
>>>Click to find more reagents for COVID-19 vaccine development and evaluation
Scenario 3: Serological diagnostic kit development
NIH used the S-2P protein for their diagnostic kit development. They used a few different methods to characterize their recombinant S trimer protein to ensure proper protein function including SDS-PAGE, SEC-MALS, and negative stain for electron microscopy (Fig. 5).
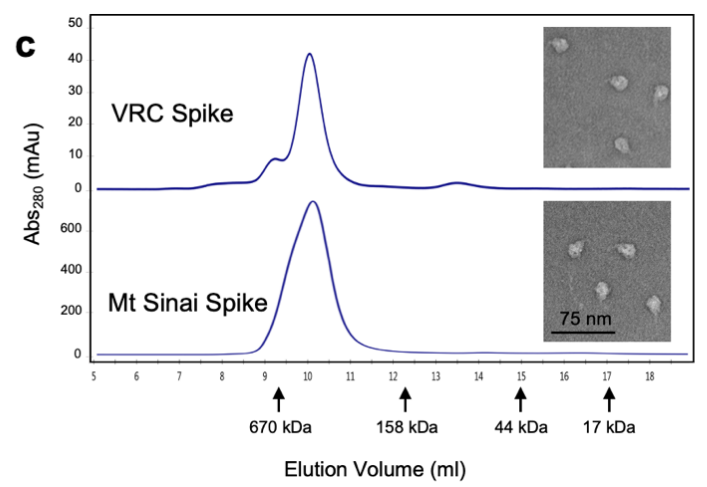
Fig. 5 Characterization of trimeric S protein
When evaluating the immunogenicity of the COVID-19 vaccine, ELISA is usually used to measure the titers of specific antibodies against the S protein. S-2P protein is widely used in these ELISA assays for antibody detection4.
>>>Click to find more reagents for COVID-19 diagnostics
Scenario 4: Neutralizing antibody screening
Currently, most of the therapeutic antibodies in the clinical stage are screened out using RBD protein. But it actually can bring you more benefit if using S-2P protein for screening. In addition to the antibodies binding to RBD, you can get antibodies that bind to NTD and S2 at the same time. These antibodies could be used in combination, and potentially will bring better therapeutic outcome (Fig. 6) 5.
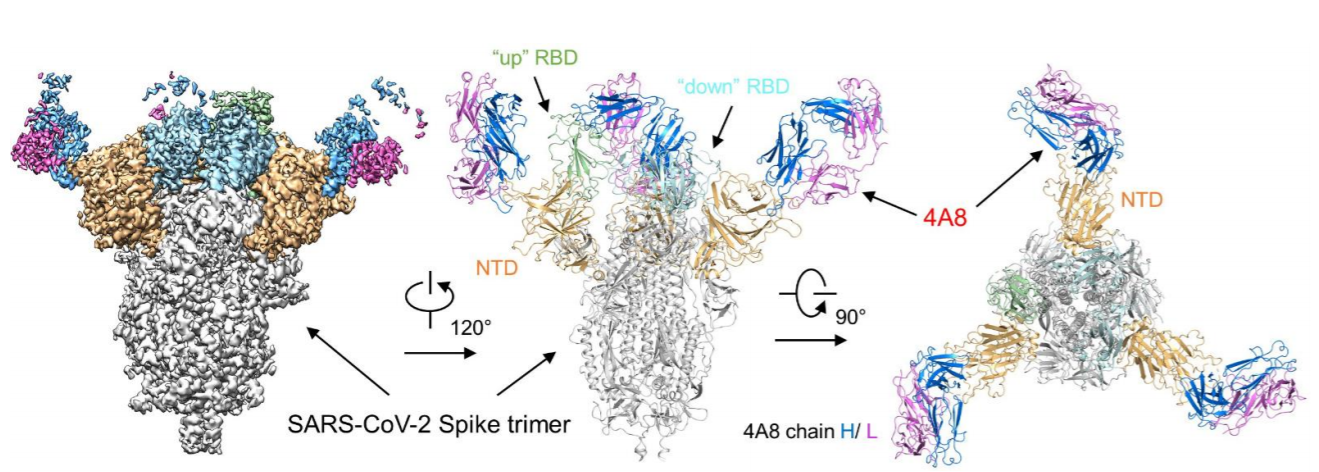
Fig. 6 Cryo-EM of S protein and NTD domain
Recently, scientists found that 4 additional proline mutations of the S2 subunit (S-6P) can further stabilize the prefusion state of the S trimer protein. Compared to S-2P, S-6P should have better performance in all the scenarios above (Fig. 7).6
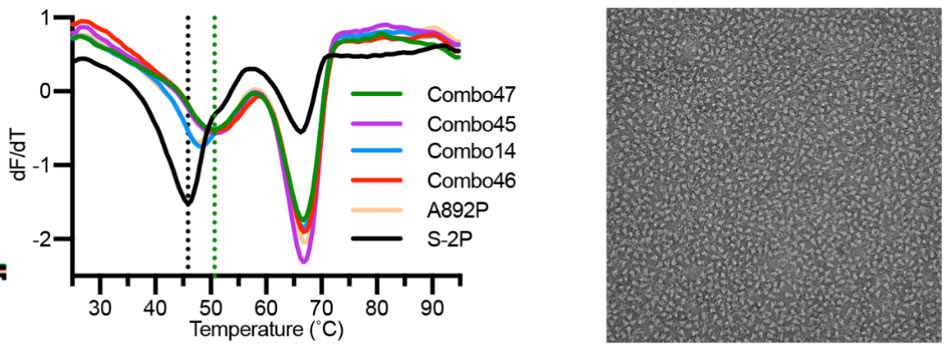
Fig.7 The stability and negative staining results of S-6P protein
>>>Click to find more SARS-CoV-2 related products
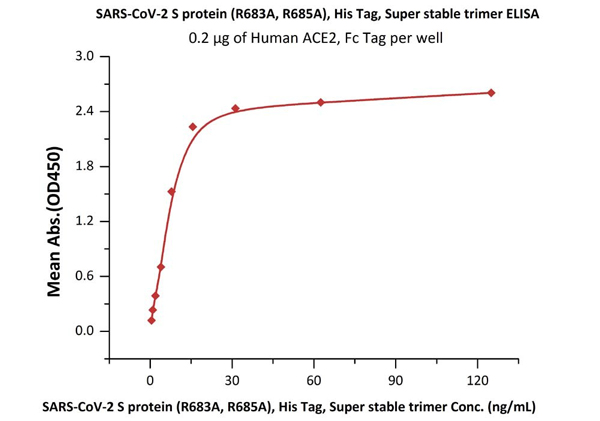
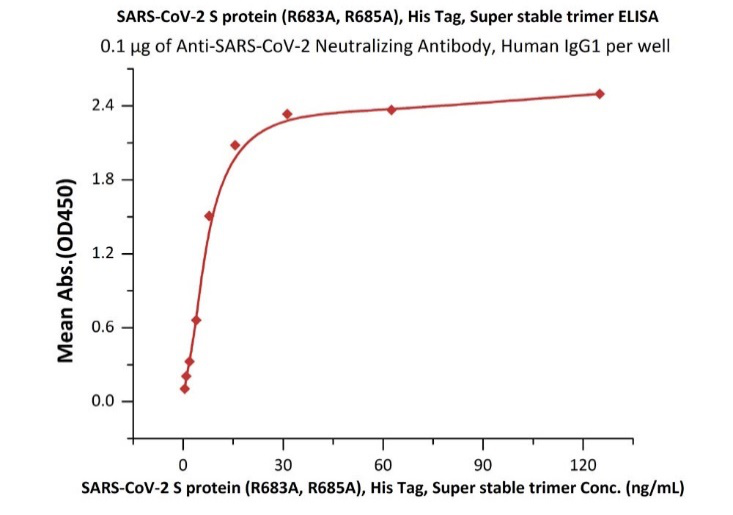
ACROBiosystems has specially designed a highly active S trimer protein. This is the only verified trimeric S protein on the market. Our SEC-MALS and negative-stain EM data reveal that ACRO's S protein is in the correct trimer form under physiological conditions, and purity is over 90%. The ELISA data shows that the S trimer protein binds to human ACE-2 protein and anti-SARS-CoV-2 neutralizing antibody well.
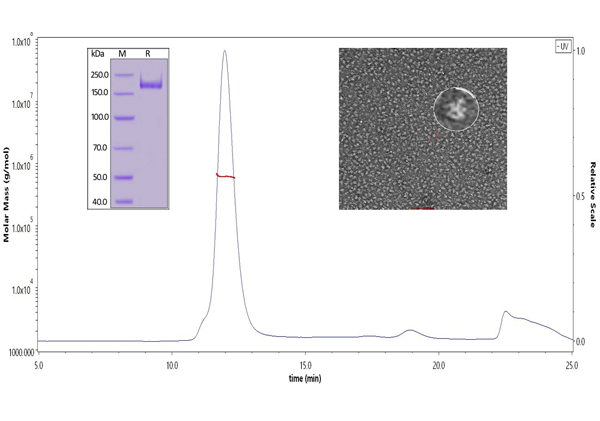
Fig 8. The purity of SARS-CoV-2 S protein, His Tag, Super stable trimer (Cat. No. SPN-C52H9) is more than 90% verified by SDS-PAGE under reducing (R) condition. The molecular weight was around 550-660 kDa confirmed by SEC-MALS. The particles are similar in size and appearance to SARS-CoV-2 trimers reported in published literature verified by negative stain electron micrography.
| Molecule | Cat. No. | Species | Tag | Host | Product Description | Order/Preorder |
|---|---|---|---|---|---|---|
| S protein | SPN-C52H3 | SARS-CoV-2 | His Tag | HEK293 | SARS-CoV-2 S protein (D614G), His Tag, Super stable trimer (MALS verified) | |
| SPN-C52H9 | SARS-CoV-2 | His Tag | HEK293 | SARS-CoV-2 S protein, His Tag, Super stable trimer (MALS & NS-EM verified) | ||
| SPN-C82E3 | SARS-CoV-2 | His Tag & Avi Tag | HEK293 | Biotinylated SARS-CoV-2 S protein (D614G), His,Avitag™, Super stable trimer (MALS verified) | ||
| SPN-C82E9 | SARS-CoV-2 | His Tag & Avi Tag | HEK293 | Biotinylated SARS-CoV-2 S protein, His,Avitag™, Super stable trimer (MALS verified) |
1. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
2. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion
4. An mRNA Vaccine against SARS-CoV-2 — Preliminary Report
5. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2
6. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes
This web search service is supported by Google Inc.