Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> Insights > How the CD3 Bispecific Antibody Reactivate Tumor Suppression in TP53-compromised Tumors? Targeted therapy is a type of cancer treatment that precisely identifies and attacks certain types of cancer cells without killing normal cells. The TP53 protein normally performs its function as an intracellular tumor suppressor, but its function is easily abolished when the gene encoding it mutates at R175H. At the same time, TP53 is rendered inaccessible as a target by therapeutic molecules, resulting in uncontrolled tumor progression.Half of all human cancers contain TP53 mutations, and in many other cancers, the function of the p53 protein is compromised. The diversity of these mutations and phenotypes presents a challenge to the development of drugs that target p53 mutant cancer cells. Besides, targeting a loss of function is difficult. These are the reasons why a TP53 targeted drug has been slow in development.
Recently, Shibin Zhou's group from Johns Hopkins University, School of Medicine, published their work of a TP53/CD3 targeting bispecific antibody. They designed a bispecific antibody that can effectively activate T cells and lysed tumor cells both in vitro and in vivo.With the successful intervention of the bispecific antibody, the inactivated TP53 recovers its tumor-suppressing role as a tumor marker on cell surface. The encouraging discovery brings a new idea for TP53 targeted therapeutic development.
The P53 gene is a tumor suppressor gene, which stops the formation of tumors. Mutations in the p53 gene may cause cancer cells to grow and spread in the body. The arginine to histidine substitution at codon 175 (R175H) is the most common TP53 mutation and is the most frequent mutation in any tumor suppressor gene. The peptide HMTEVVRHC derived from the p53 R175H mutation, can bind to a particular HLA allele (HLA-A*02:01) and form a peptide HLA complex on the cell surface. Peptide-HLA (pHLA) complexes are the natural ligands for TCRs.
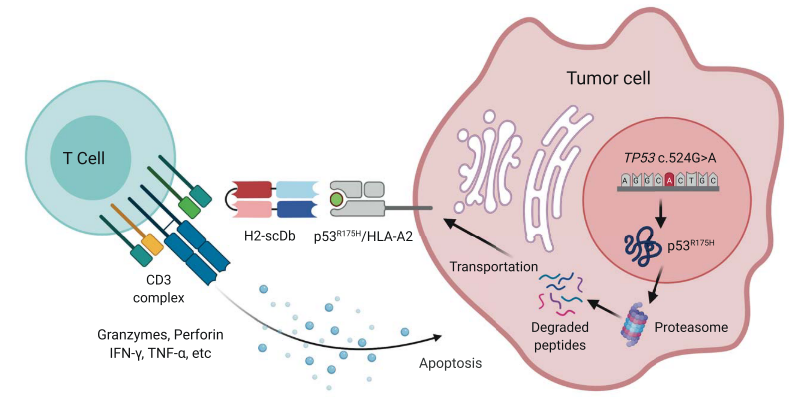
Figure 1 Illustration depicting the mechanism of action of the TP53(R175H)/CD3 bispecific antibody H2-scDb.
To identify TCRm single-chain variable fragments (scFvs) that selectively target mutant pHLA complexes, Shibin Zhou’s group screened an scFv-displaying phage library. Selected phage clones were amplified and assessed using flow cytometry. After some further selection, a few clones were converted to T cell retargeting bispecific antibodies in the format of single-chain diabody (scDb) (Figure 2). After the potency and specificity evaluation, a clone called H2-scDb was chosen for further analysis.
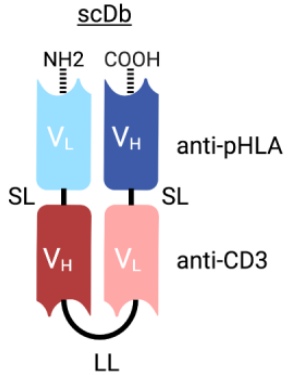
Figure 2 Structure of TP53 Targeting scDb
H2-scDb can effectively activate T cells to release granzyme, perforin, and cytokines in vivo at low concentrations, and mediate the killing of tumor cells (Figure 3).
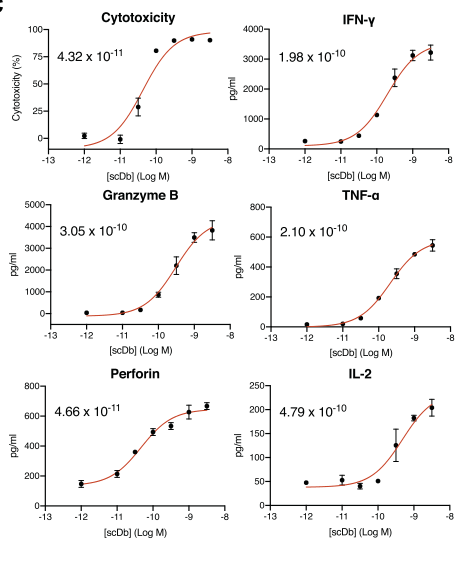
Figure 3 In vitro anti-tumor activity of H2-scDb
H2-scDb can significantly inhibit the tumor growth of xenograft mice models. While the growth of the tumor is not affected in the P53 knockout models. (Figure 4)
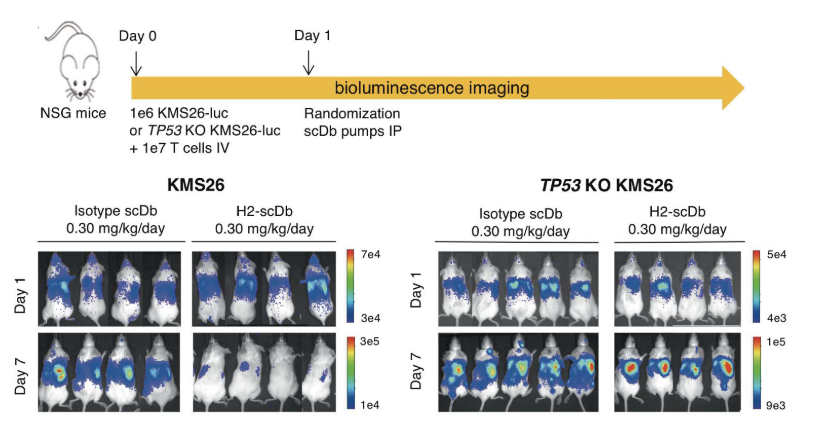
Figure 4 In vivo anti-tumor activity of H2-scDb
To understand the structural basis for the high specificity of the H2 clone for p53R175H/HLA-A*02:01, the scientists first converted H2 into a full-length immunoglobulin G (IgG) (H2-IgG) and confirmed that binding specificity was preserved in this format. The H2-IgG was then digested into an antigen-binding fragment (H2-Fab) with papain. The H2-Fab–p53R175H/HLA-A*02:01 complex was purified, and its crystal structure was determined through molecular replacement and refined to 3.5 Å resolution. The p53R175H peptide (HMTEVVRHC) occupied the binding cleft a1-a2 of HLA-A*02:01, burying a solvent-accessible surface area. The recognition of the HLA-A*02:01 by the H2-Fab was mediated by all six CDRs. By contrast, only four of the six H2-Fab CDRs (H1, H2, H3, and L3) interacted with the p53R175H peptide.
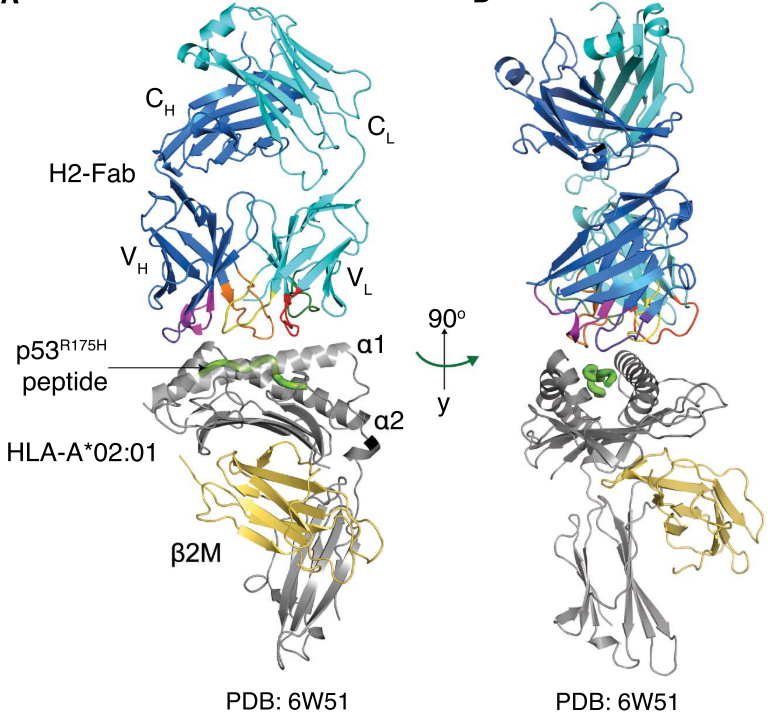
Figure 5 Crystal structure of p53R175H/HLA-A*02:01 bound to the H2-Fab fragment
This study revealed that the TP53 R175H targeting bispecific H2-scDb specifically recognizes and activates T cells. It can play an anti-tumor role while ensuring safety. They got a promising result of H2-scDb in animal studies. We look forward to the further clinical evaluation of this antibody.
Conventional antibody therapy can only target antigens on the cell surface. However, most of the tumor targets are intracellular, which makes antibody development extremely difficult. In this study, the H2-scDb antibody was developed to recognize the antigen peptide/MHC complex presented on the surface of tumor cells and activate the anti-tumor effect of T cells.
This successful story provides new ideas and supports for antibody development against similar targets and solid tumor CAR-T cell therapy development.
In this study, the scientist used ACROBiosystems’ CD3E & CD3D heterodimer protein (Cat.No. CDD-H82W6) for the binding affinity study of scDb.
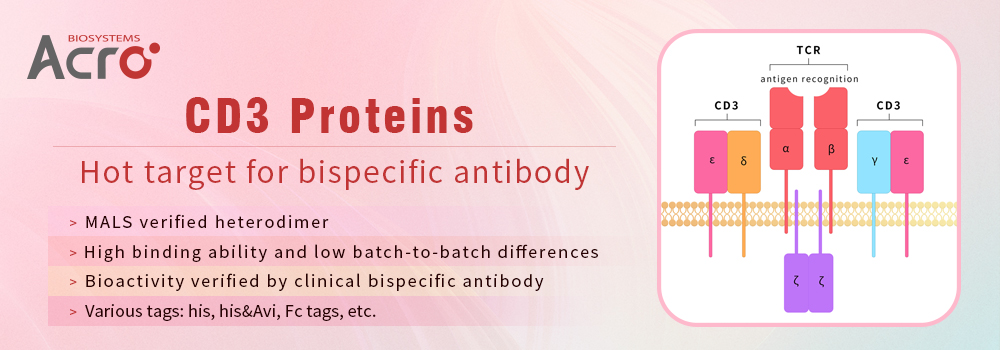
ACROBiosystems has developed a series of homogeneous CD3δ/CD3ε and CD3γ/CD3ε proteins with good bioactivity. They are verified as 1:1 heterodimers by nonreductive electrophoresis and MALS as well. Additionally, they show very good performance in therapeutic antibody screening, identification, characterization as well as clinical pharmacokinetic assays, etc. What’s more, our CD3 products were successfully used for the clinical development of a bispecific antibody, which greatly accelerated the process of therapeutic antibody development.
ACROBiosystems has developed a series of homogeneous CD3 proteins with good bioactivity, which are verified as 1:1 heterodimer by nonreductive electrophoresis and MALS.
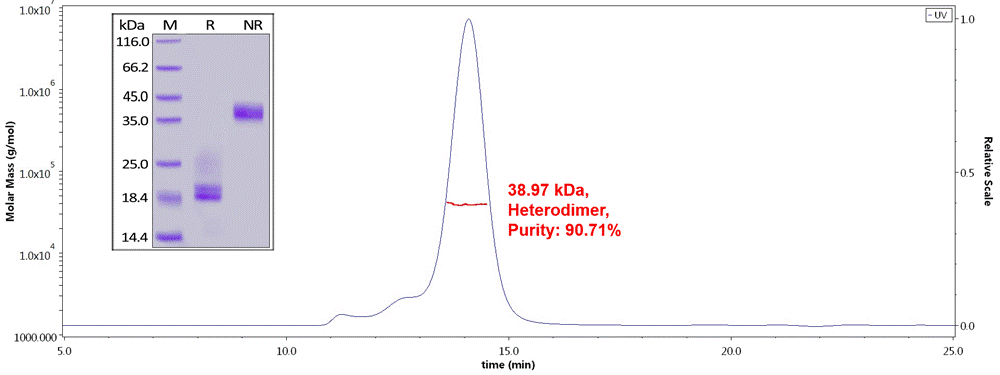
Fig.1 Human CD3E&CD3D Heterodimer Protein, His Tag&Tag Free (Cat. No. CDD-H52W1) on SDS-PAGE under reducing (R) and non-reducing(NR)condition. The purity of the protein is more than 85% and around 35-43 kDa verified by SEC-MALS.
These verified authentic 1:1 heterodimer proteins show high bioactivity in different applications. The products are homogenous with strict quality control to guarantee the minimal batch-to-batch differences.
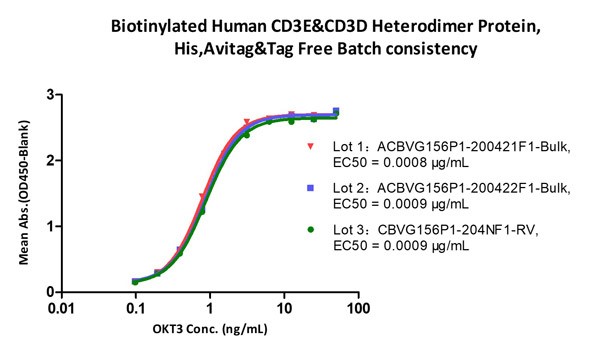
Fig.2 Immobilized Biotinylated Human CD3E&CD3D Heterodimer Protein, His,Avitag&Tag Free (Cat. No. CDD-H82W6) at 1 μg/mL (100 μL/well) on Streptavidin (Cat. No. STN-N5116)precoated (0.5 μg/well) plate, can bind Monoclonal Anti-Human CD3 Antibody, Mouse IgG2a (Clone: OKT3) (Cat. No. CDE-M120a) with a linear range of 0.2-6 ng/mL (QC tested), and batch differences EC50<0.0001 μg/mL.
These products are suitable for various applications at different stages of drug development, such as antibody immunotiter detection, biopanning, antibody screening, affinity characterization and clinical pharmacokinetic assays.
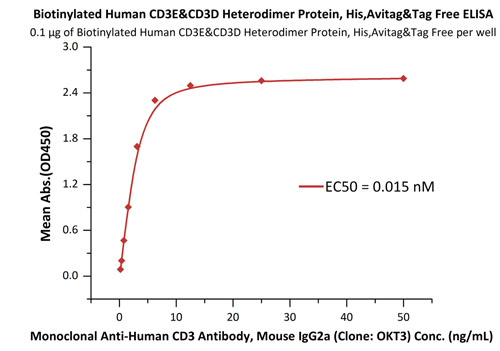
Fig.3 Immobilized Biotinylated Human CD3E&CD3D Heterodimer Protein, His,Avitag&Tag Free (Cat. No. CDD-H82W6) at 1 μg/mL (100 μL/well) on streptavidin precoated (0.5 μg/well) plate, can bind Monoclonal Anti-Human CD3 Antibody, Mouse IgG2a (Clone: OKT3) (Cat. No. CDE-M120a) with a linear range of 0.2-6 ng/mL.
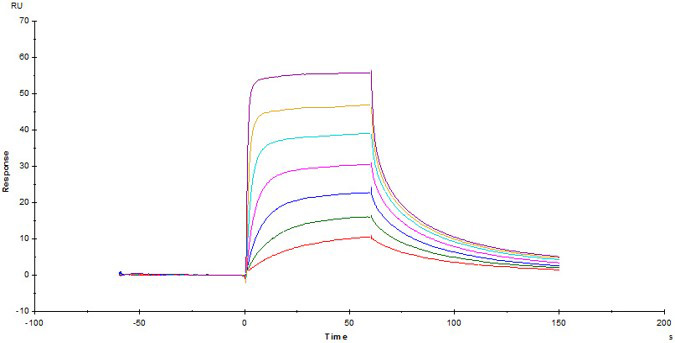
Fig.4 Biotinylated Human CD3E&CD3D Heterodimer Protein, His,Avitag&Tag Free (Cat. No. CDD-H82W6) captured on Biotin CAP - Series S sensor Chip can bind Anti-Human CD3 Antibody, Mouse IgG2a (Clone: OKT3) with an affinity constant of 22.5 nM as determined in a SPR assay (Biacore T200) .
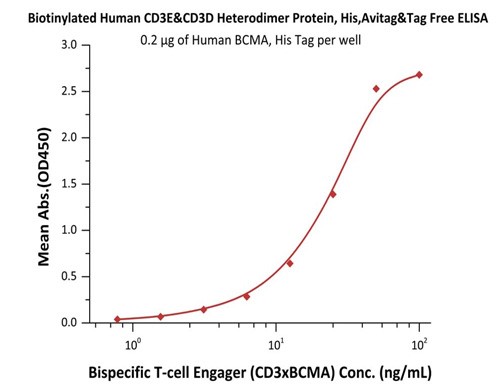
Fig.5 Immobilized Human BCMA, His Tag (Cat. No. BCA-H522y) at 2 μg/mL, add increasing concentrations of Bispecific T cell Engager (CD3 X BCMA) in 10% human serum and then add Biotinylated Human CD3E&CD3D Heterodimer Protein, His,Avitag&Tag Free (Cat. No. CDD-H82W6) at 0.2 μg/mL. Detection was performed using HRP-conjugated streptavidin with sensitivity of 15 ng/mL.
Reference:
Emily Han-Chung Hsiue et al. Targeting a neoantigen derived from a common TP53 mutation. Science, 2021, doi: 10.1126/science.abc8697.
This web search service is supported by Google Inc.