Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
| Cat . Non | Espèces | Description du produit | Structure | Pureté | Caractéristique |
|---|---|---|---|---|---|
| CAS-C001 | Streptococcus pyogenes | resDetect™ Cas9 (CRISPR Associated Protein 9) ELISA Kit (Residue Testing) | |||
| GMP-CA9S18 | Streptococcus pyogenes | GMP GENPower™ NLS-Cas9 Nuclease |
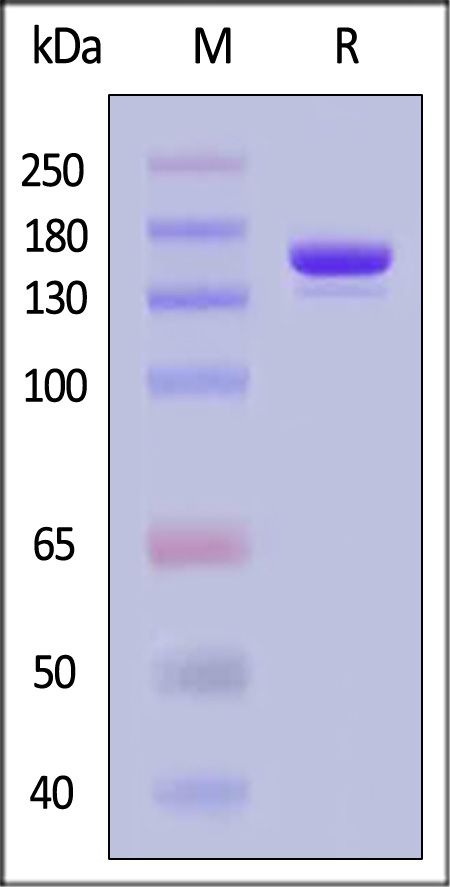
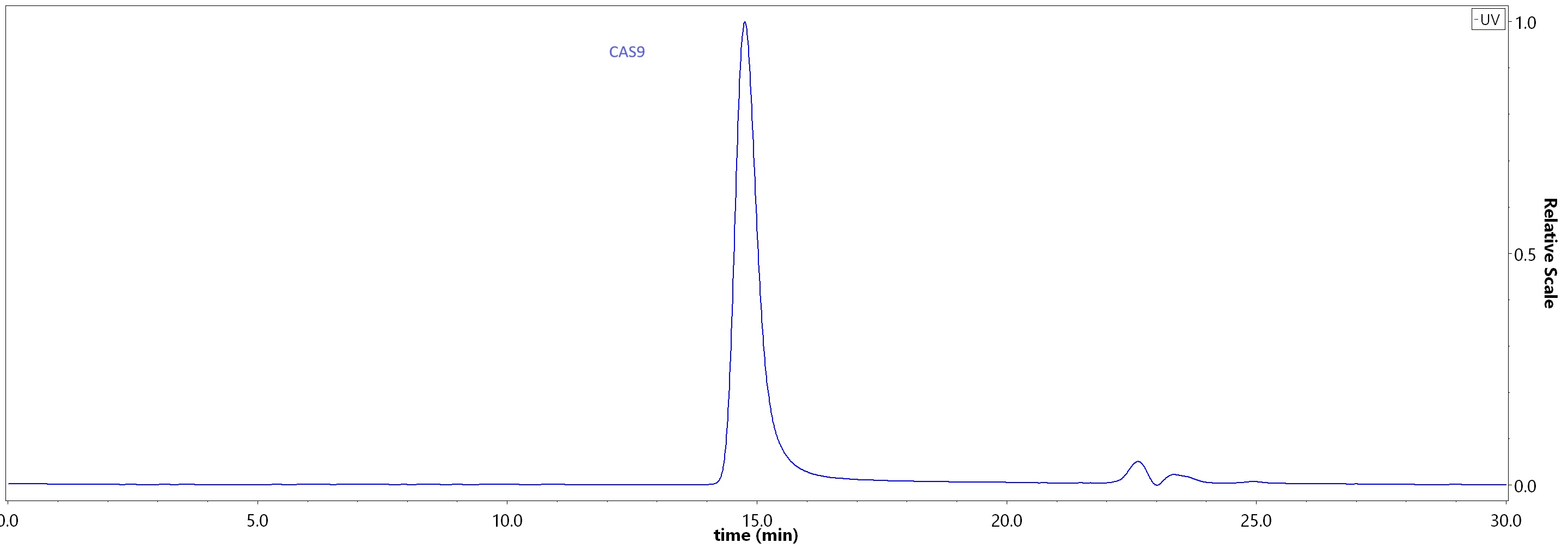
|
||
| CA9-S5149 | Streptococcus pyogenes | GENPower™ NLS-Cas9 Nuclease, premium grade |
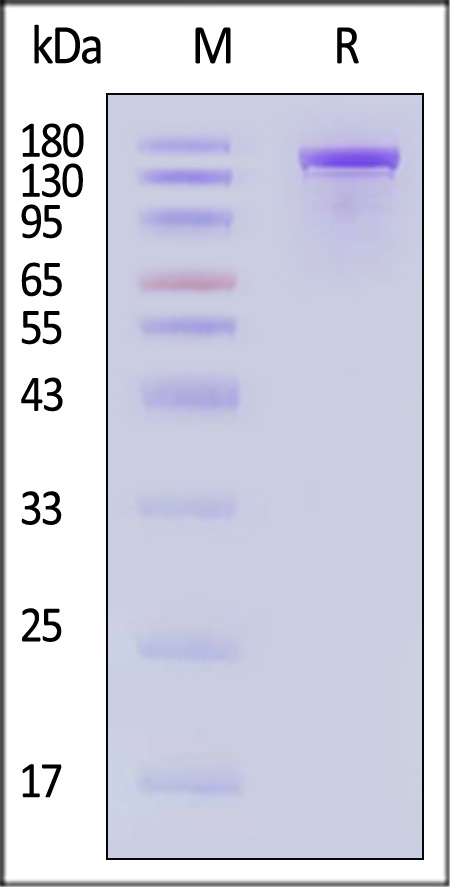
|
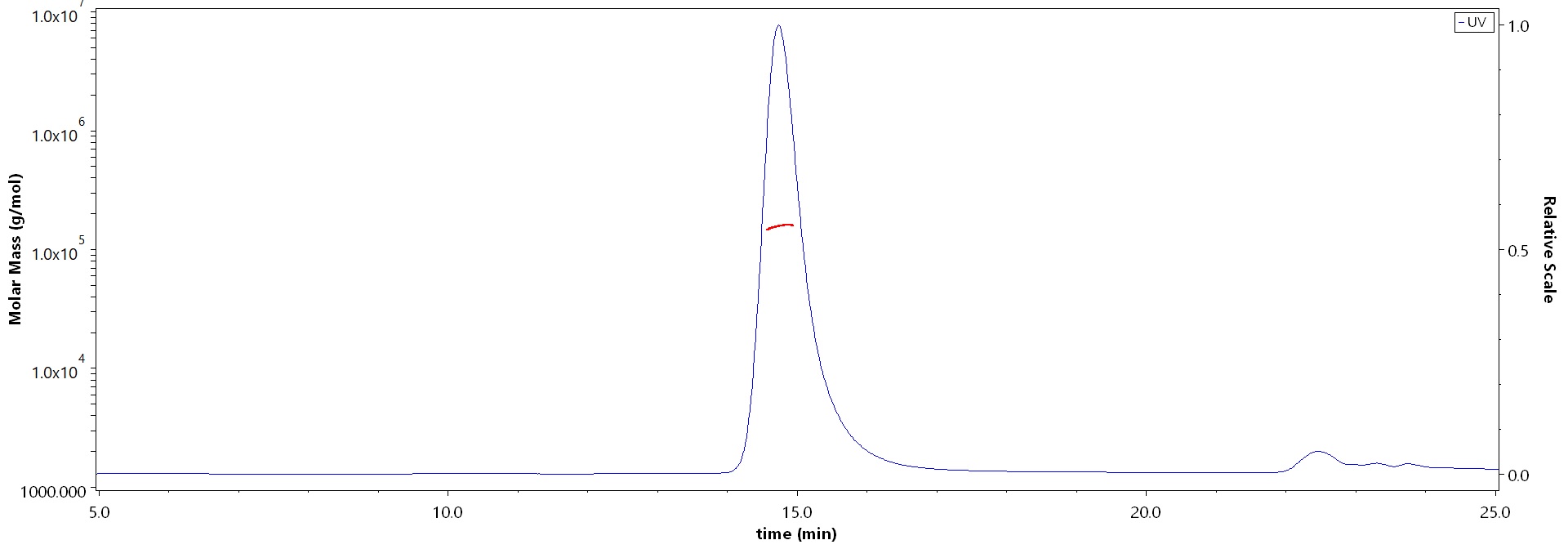
The purity of GENPower™ NLS-Cas9 Nuclease, premium grade (Cat. No. CA9-S5149) is more than 95% and the molecular weight of this protein is around 140-180 kDa verified by SEC-MALS.
This web search service is supported by Google Inc.








