Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>































 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
SARS-CoV-2 is changing slowly as it spreads. At the 614th amino-acid position of the spike protein, the amino acid aspartate (D, in biochemical shorthand) was regularly being replaced by glycine (G). Virologists call it the D614G mutation. This mutant occupies an increasing proportion of detected variants since the end of February 2020, which has made it the most commonly identified variant in many parts of the world at present. This attracted researchers’ attention to study if the D614G mutation is more infectious/transmissible or more deadly?
Recently, a research article titled “Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant” was published in Cell. Scientists studied the D614G mutation from different aspects of cell biology and structural biology. We will walk you through a few highlights of this article below.
According to the GISAID SARS-CoV-2 database as of June 25, 2020, the global frequency of the S protein D614G variant has increased steadily over time and is now present in approximately 74% of all published variants. (Fig. 1)
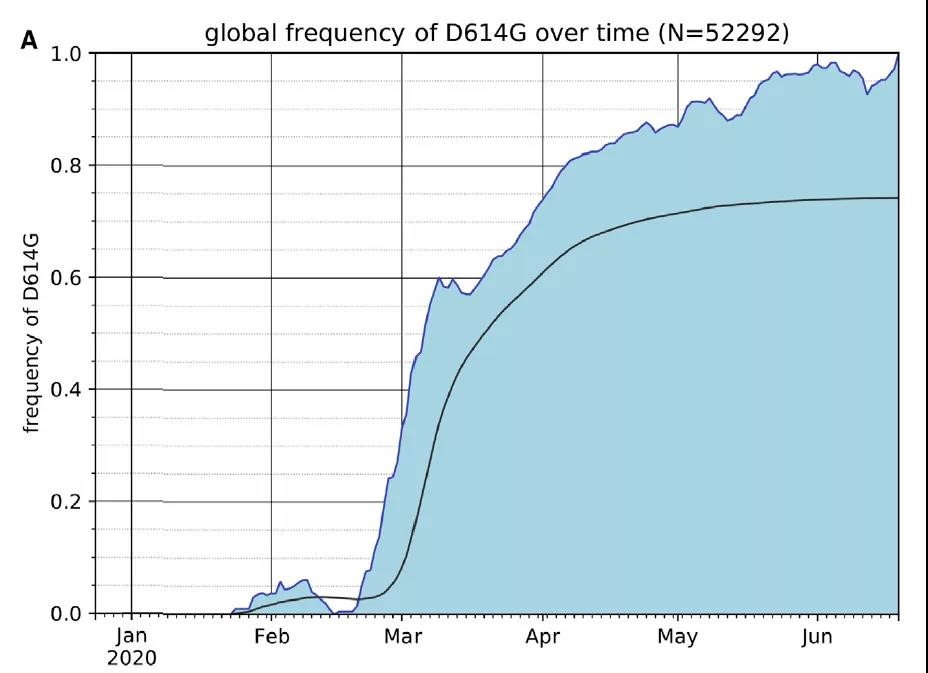
Fig. 1 The Frequency of the SARS-CoV-2 S Protein D614G Variant over the Course of the Pandemic Has Increased Nearly to Fixation
To compare the ability of the ancestral S protein D614 or the D614G variant to target virions for infection, they constructed the pseudotyped lentivirus. A few different cell models are studied including Calu-3 human lung epithelial cells, Caco-2 human colon epithelial cells, HEK293 cells, and SupT1 cells. According to the data in Fig. 2, the infectivity of D614G is 4 to 9 fold higher than D614.
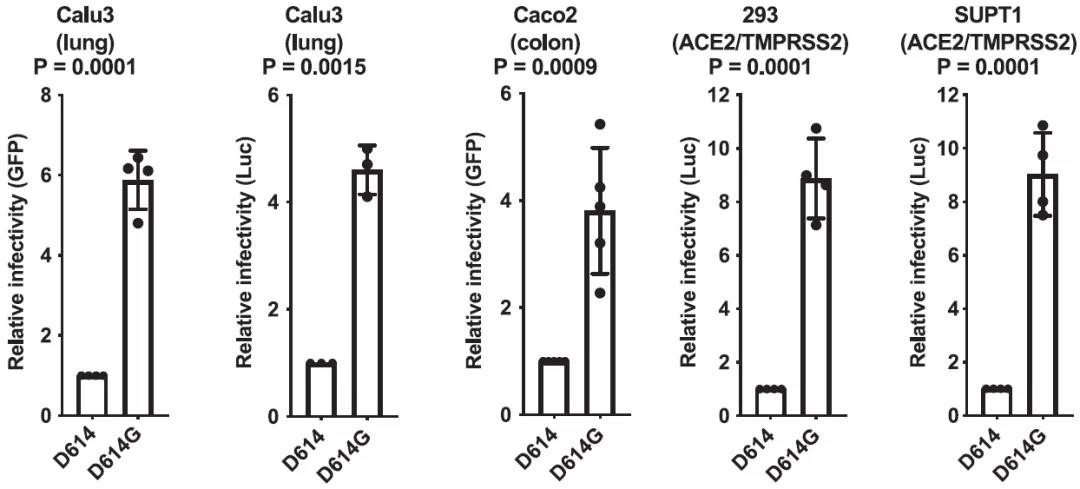
Fig. 2 SARS-CoV-2 D614G S Protein Variant Enhances Infectivity of Pseudotyped Lentiviruses in Cell Culture.
For SARS-CoV-2 to enter human cells, the spike protein on the surface of the virus must bind to the host receptor protein, ACE2. Although D614G is located outside of the receptor-binding domain, this mutation might alter ACE2-binding properties via allosteric effects. According to the surface plasmon resonance (SPR) results in Fig. 3, D614G decreases the affinity for ACE2 by increasing the rate of dissociation. These data demonstrated that the increased infectivity of D614G is not explained by greater ACE2 binding strength.
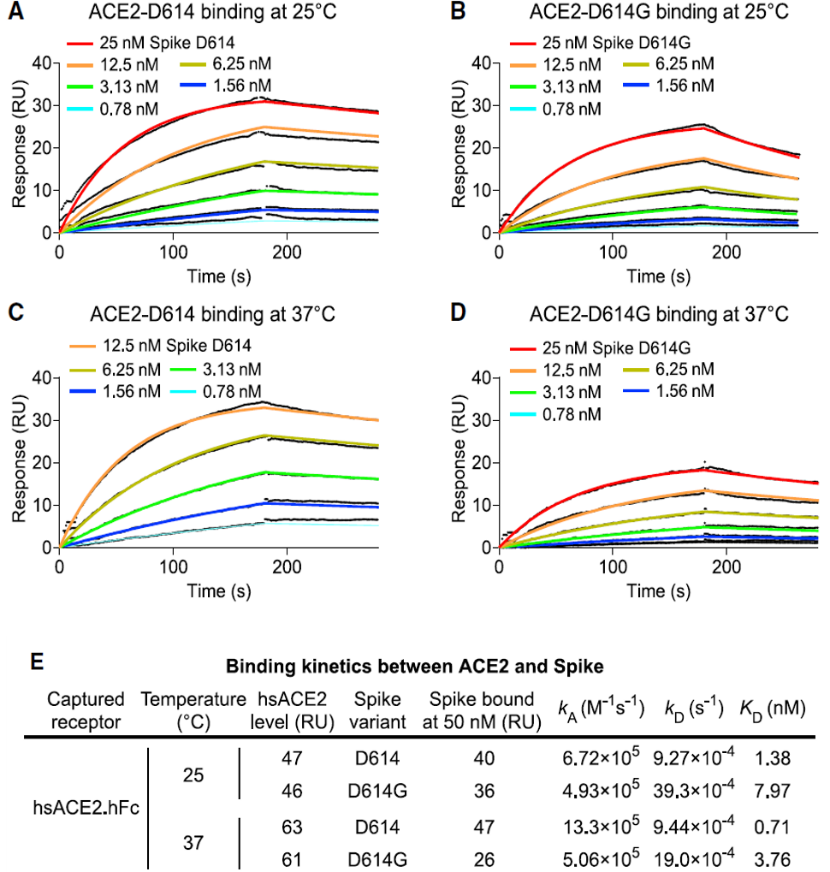
Fig. 3 SARS-CoV-2 D614G S Protein Variant Binds ACE2 Weaker than the Ancestral Protein.
So far, most of the COVID-19 therapeutic antibodies are developed targeting the receptor-binding domain of spike protein. Although this structural change is non-conservative, the question is still raised if it will compromise the effectiveness of these therapeutic antibodies currently under development. They did some studies using Regeneron antibodies. The results are summarized below in Fig. 4, suggesting D614G and the ancestral D614 are equally sensitive to these neutralizing antibodies targeting the receptor-binding domain.
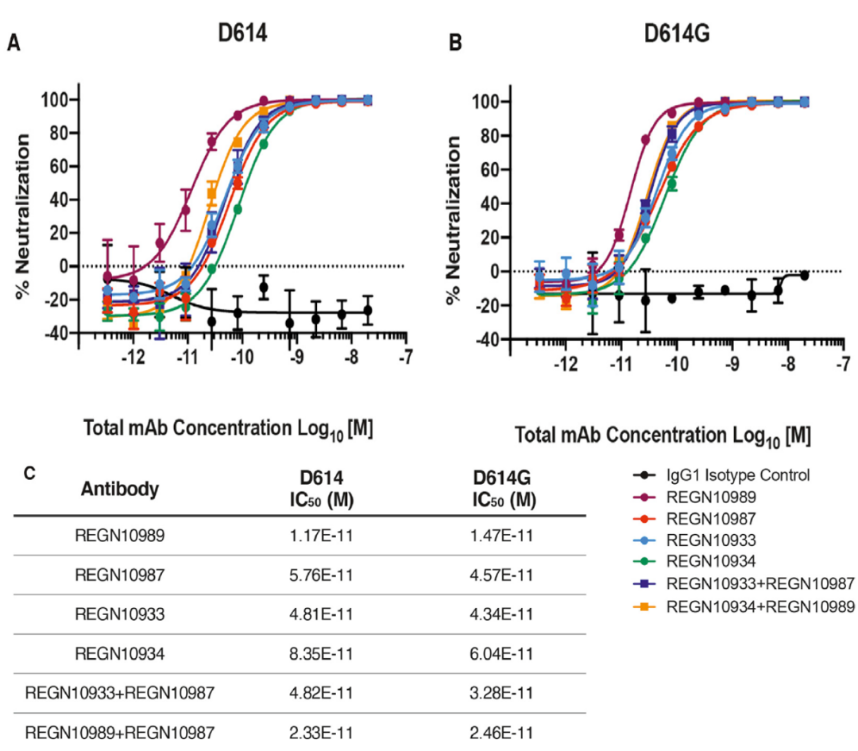
Fig. 4 Neutralization Potency of Monoclonal Antibodies Targeting the SARS-CoV-2 S Protein Receptor-Binding Domain Is Not Attenuated by D614G.
There are mainly two conformational states for SARS-CoV-2 spike trimer protein, the “up” representing a receptor-accessible state and the “down” representing a receptor-inaccessible state (Fig. 5). The “up” state accounts for 54% while the “down” state accounts for 46%.[2]
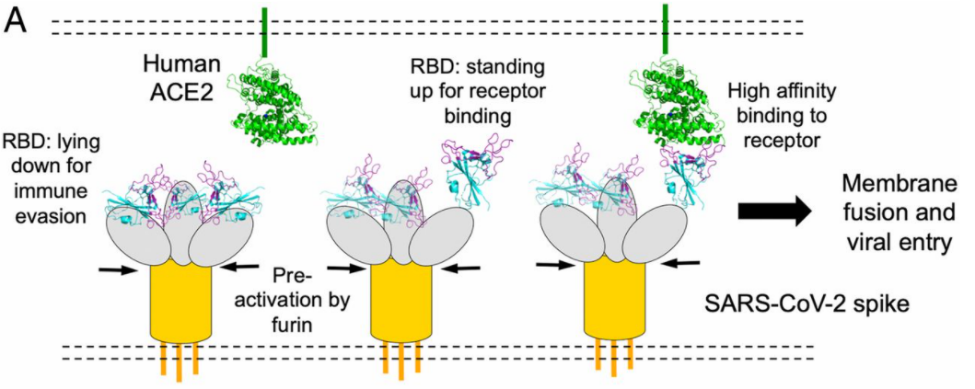
Fig. 5 Spike protein receptor binding domain binding to ACE2 for viral entry .[3]
To further study if there is any conformation change caused by D614G mutation, scientists did Cryo-EM testing. They found that there was no significant change in the S2 subunit. But there was a shift in the D614G S1 subunit, causing the following receptor binding domain conformation change (Fig. 6). As we can see, there is a total of 95% “up” state conformation in D614G, which significantly increases the chance of infection.
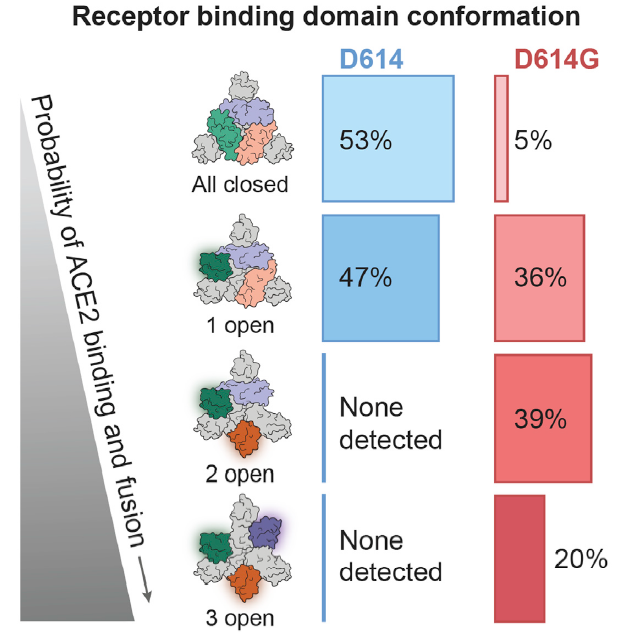
Fig. 6 D614G mutation increased the amount of RBD 2 open and 3 open.
Currently, almost 200 COVID-19 vaccine development projects are going on. When evaluating the efficacy of the vaccine, scientists usually use spike protein or receptor binding domains to measure the antibody titer. It is worth mentioning that Moderna used D614G spike protein instead in these assays.[5]
Trimeric Spike protein (D614G) products from ACROBiosystems
ACROBiosystems developed a few stable recombinant spike trimer protein products including D614G spike trimer protein, ancestral D614G spike trimer protein as well as related protein-coupled magnetic beads, which are all designed for COVID-19 therapeutic and vaccine development.
Assay Data
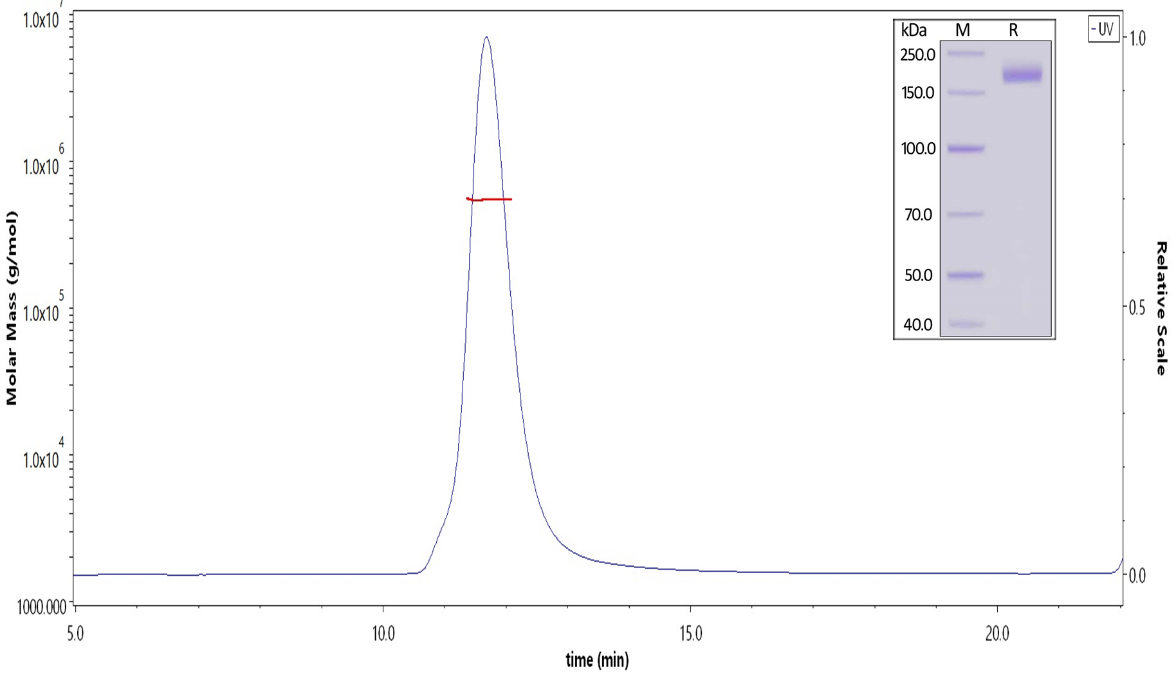
Fig 1. The purity of SARS-CoV-2 S protein (D614G), His Tag, Super stable trimer (Cat. No. SPN-C52H3) is more than 90% and the molecular weight of this protein is around 520-620 kDa verified by SEC-MALS.
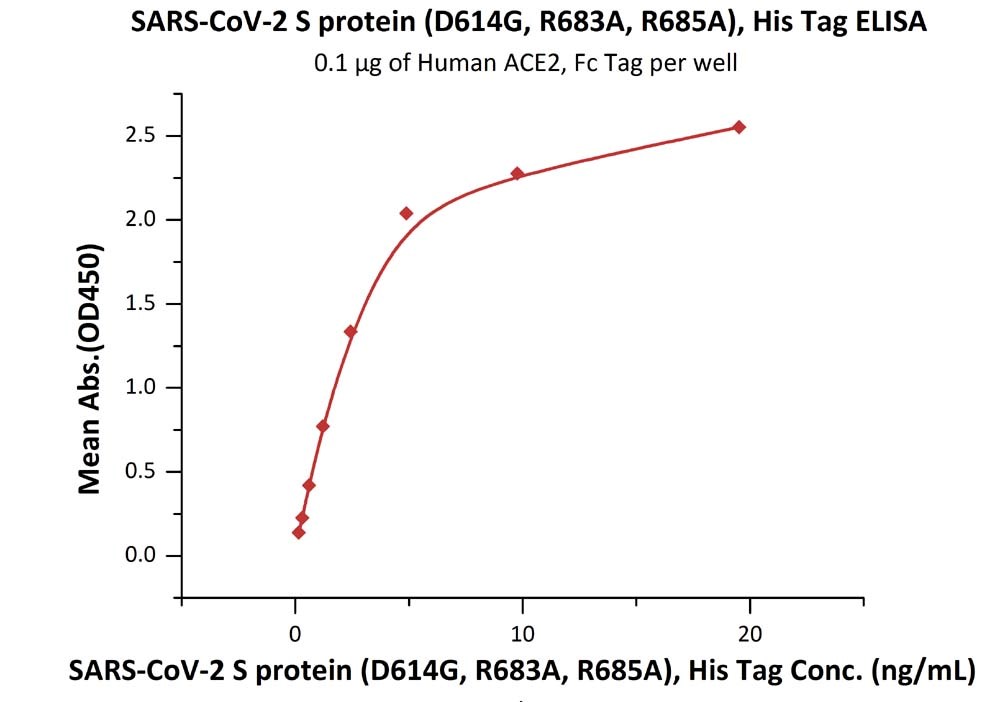
Fig 2. Immobilized Human ACE2, Fc Tag (Cat. No. AC2-H5257) at 1 μg/mL (100 μL/well) can bind SARS-CoV-2 S protein (D614G), His Tag, Super stable trimer (Cat. No. SPN-C52H3) with a linear range of 0.2-5 ng/mL.
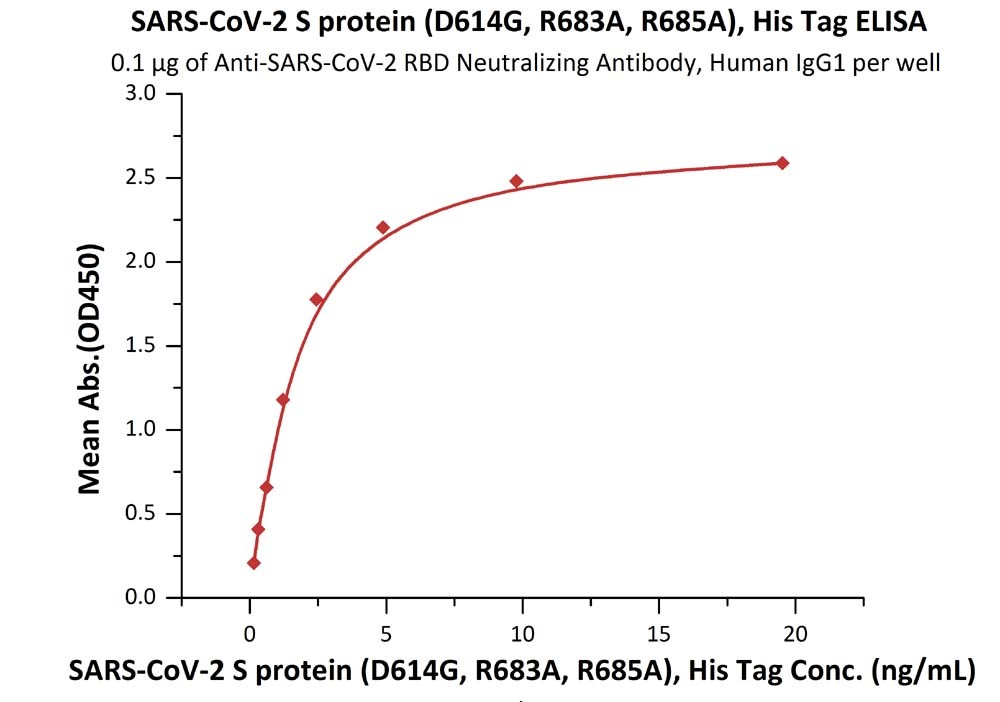
Fig 3. Immobilized Anti-SARS-CoV-2 RBD Neutralizing Antibody, Human IgG1 (Cat. No. SAD-S35) at 1 μg/mL (100 μL/well) can bind SARS-CoV-2 S protein (D614G), His Tag, Super stable trimer (Cat. No. SPN-C52H3) with a linear range of 0.2-2 ng/mL.
References:
1. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes.
2. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation.
3. Cell entry mechanisms of SARS-CoV-2 .
4. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy.
5. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults.
This web search service is supported by Google Inc.







