Leave message
Can’t find what you’re looking for?
Fill out this form to inquire about our custom protein services!
Inquire about our Custom Services >>






























 Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.  Limited Edition Golden Llama is here! Check out how you can get one.
Limited Edition Golden Llama is here! Check out how you can get one.
 Offering SPR-BLI Services - Proteins provided for free!
Offering SPR-BLI Services - Proteins provided for free!  Get your ComboX free sample to test now!
Get your ComboX free sample to test now!
 Time Limited Offer: Welcome Gift for New Customers !
Time Limited Offer: Welcome Gift for New Customers !  Shipping Price Reduction for EU Regions
Shipping Price Reduction for EU Regions
> Insights > Large-scale Characterization of Drug Candidates Against Transmembrane Receptors using HT-SPR Membrane targets make up a substantial part of the overall “undruggable” therapeutic space that has recently garnered widespread interest. Despite encouraging improvements in the tools to screen for therapeutics against membrane-bound targets, there are still many practical limitations owing to the challenges of working with proteins that are not highly stable outside of the cell-membrane environment. Furthermore, the use of high-quality, full-length transmembrane receptors is critical for accurate characterization of drug candidates. High-throughput surface plasmon resonance (HT-SPR) is a powerful technique that is transforming characterization workflows and enabling a greater breadth and depth of information for up to thousands of drug candidates. Here we demonstrate the ability to quantitatively assess binding kinetics for panels of antibodies against full-length transmembrane receptors in several formats. This workflow highlights opportunities to perform detailed binding characterization for up to thousands of drug candidates in parallel, accelerating the discovery of drug candidates targeting transmembrane receptors.
Figure 1 illustrates the three membrane receptor formats tested in these studies: virus-like particle (VLP), Nanodisc, and detergent solubilized. All membrane receptor formats tested were from ACROBiosystems. GPRC5D was tested in the following formats: VLP (#GPD-H52P7), Nanodisc (#GPD-H52D4), and detergent solubilized (#GPD-H52D3). CCR5 receptor was tested as VLP (#CC5-H52P3) and detergent solubilized (#CC5-H52D1). For the ELISA studies, anti-GPRC5D antibody was used for all membrane receptor formats. Structural studies using dynamic light scattering was performed to verify the consistency of VLP size. Affinities and binding kinetics of mAbs against the three membrane receptor formats were ascertained by using HT-SPR. For VLP studies the Carterra LSA instrument was primed with HBS + 0.005% Tween-20 + 5% trehalose, pH 7.4, for Nanodisc HBS + 0.005% Tween-20 + 5% glycerol, pH 7.4, and for detergent solubilized HBS + 5% glycerol + 0.05%DDM/0.01%CHS detergent (ACROBiosystems #DC-11), pH 7.4. Sample deck temperature was 20°C and the interaction surface temperature was 25°C. Antibodies against GPRC5D and CCR5 were captured onto a Protein A/G sensor chip (Carterra #4292) at 1ug/ml. For the GPRC5D studies, mAb clone 571961R (R&D Systems) was tested. For CCR5 a proprietary panel of 72 mAbs was used. Either single concentration (VLP studies) or ascending titrations (two-fold concentrations up to 250 nM for Nanodisc and detergent-solubilized studies) of each receptor were injected across the arrays. At the end of the injections the array was regenerated with 50mM glycine pH 2.0 and the array was captured with fresh mAbs in preparation for binding with the next receptor format.
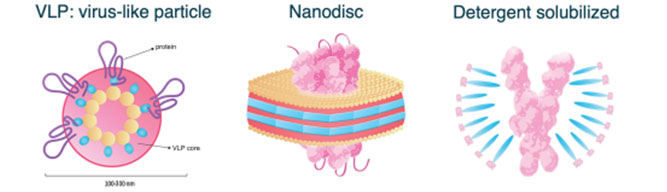
Figure 1. Three membrane receptor formats used for large-scale characterization. Adapted from www.acrobiosystems.com.
Structural characterization was performed for all three membrane receptor formats of GPCR5D. Since VLP particles consist of multiple membrane-bound receptors, dynamic light scattering (DLS) was performed to evaluate particle size, revealing a mean peak radius of 65-85 nm. For Nanodisc and detergent solubilized receptors, SDS-PAGE was conducted to confirm structure and purity (>90%).
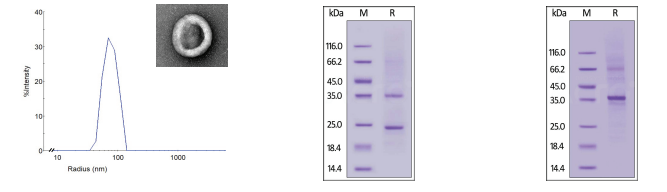
Figure 2. (Left) DLS evaluation of VLP GPCR5D with an electron microscopy images to confirm VLP shape and size. (Center, Right) SDS-PAGE evaluation of Nanodisc and detergent micelle GPCR5D, respectively.
Recombinant protein bioactivity was tested using ELISA. Human IgG4 anti-GPCR5D antibodies were used to test the bioactivity of GPCR5D in each format. The resulting linear ranges varied significantly depending on the format with VLP at 0.2-8 ng/mL, Nanodisc at 10 – 80 ng/mL, and 2 – 78 ng/mL for detergent micelle.
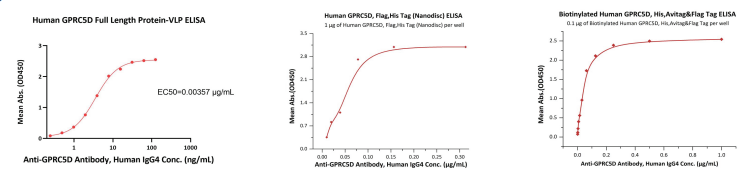
Figure 3. ELISA assays were performed on (Left) VLP, (Center) Nanodisc, (Right) Detergent Micelle GPCR5D
Example binding responses for one anti-GPRC5D mAb are shown in Figure 2. While the Nanodisc and detergent-solubilized forms have similar kinetic profiles, the VLP format demonstrates the expected avidity profile with much slower dissociation. The lack of obvious avidity in the GPRC5D Nanodisc format, due to its consistency with the detergent solubilized binding profile, suggests a single copy number in each Nanodisc on average. In all three cases, the binding is robust suggesting any one of these formats would be appropriate for screening large panels of therapeutic candidates to identify valid binders.
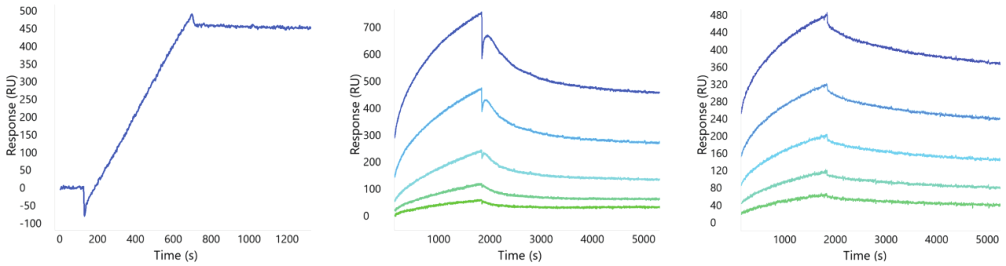
Figure 4. Binding profiles of GPRC5D in VLP, Nanodisc, and detergent solubilized formats.
Representative sensorgram profiles of mAbs binding to CCR5 formatted as a VLP are shown in Figure 5. Very slow dissociation profiles indicate avidity-driven kinetics, similar to what was observed for VLP formatted GPRC5D. Specificity of the assay is highlighted by lack of binding against certain clones in the panel, indicated in gray. In Figure 6, the sensorgram profiles for the full panel of 72 anti-CCR5 mAbs are shown, indicating a diversity of kinetic profiles against the detergent solubilized form of CCR5. A zoomed view for six of these mAbs is shown in Figure 7, with a 1:1 Langmuir (top) or heterogeneous (bottom) model fit lines in red overlaid on the sensorgrams. The data fit well to the model, indicating that detergent solubilized receptors provide a viable path to determining binding kinetics for panels of therapeutic candidates.
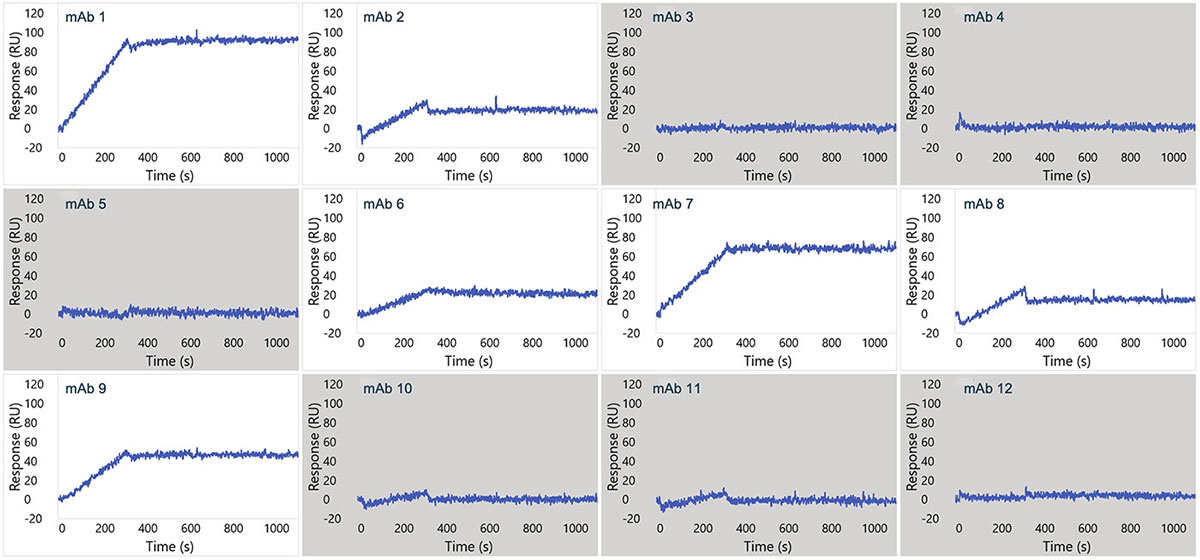
Figure 5. Single concentration sensorgrams for 12 mAbs showing binding to the VLP form of CCR5. Gray boxes indicate no detectable binding.
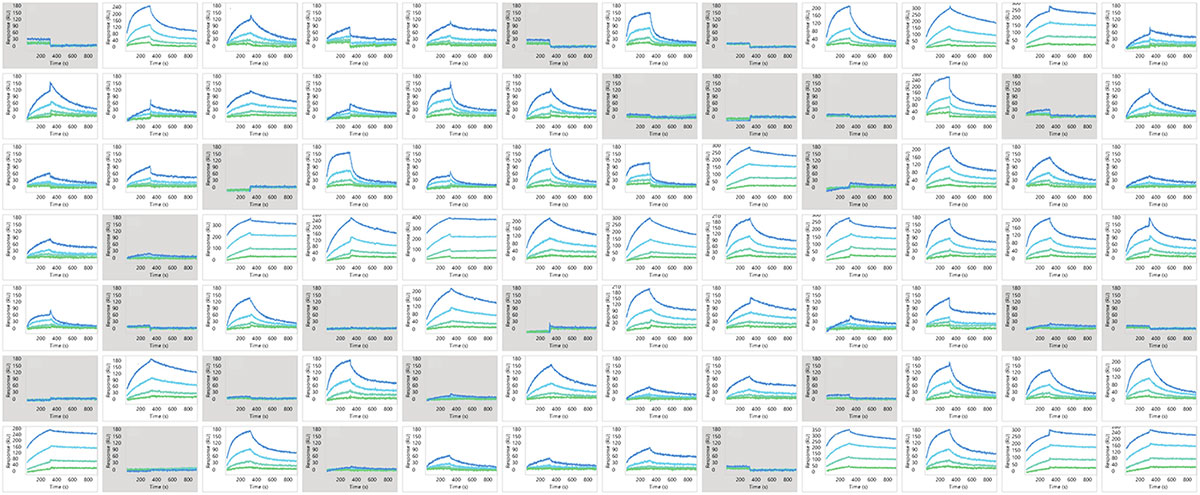
Figure 6. Binding profiles of 72 mAbs against detergent solubilized CCR5 receptor. Gray boxes indicate no detectable binding.
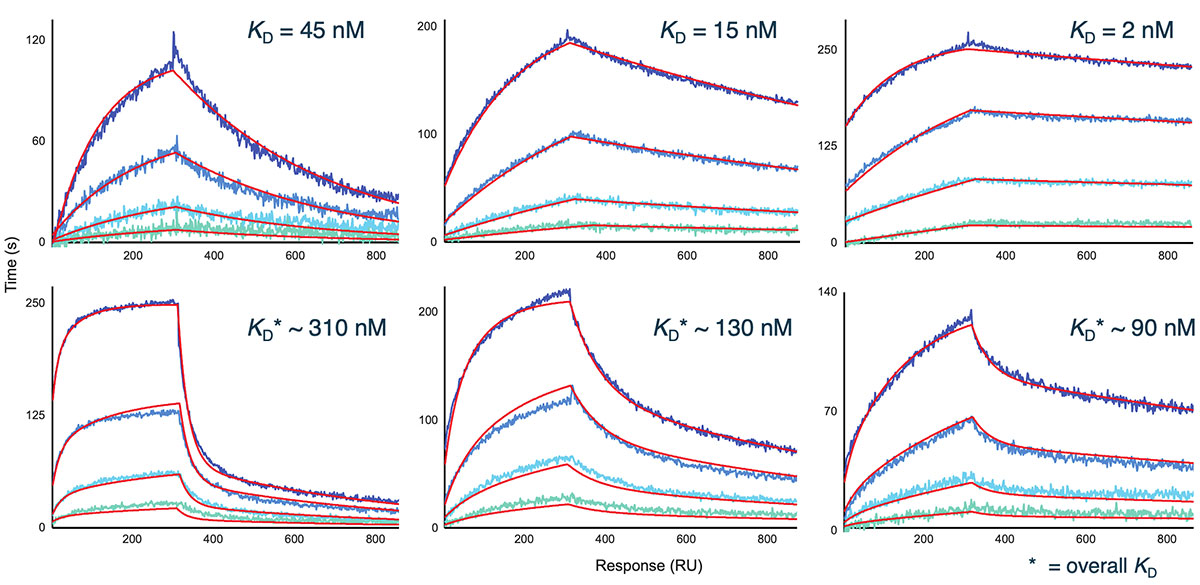
igure 7. Representative 1:1 Langmuir (top) and heterogeneous (bottom) fits (red lines) for mAbs against detergent-solubilized CCR5 receptor.
Paired with high-quality full-length transmembrane receptors from ACROBiosystems, HT-SPR analysis using Carterra’s LSA instrumentation enables large-scale characterization of potential drug targets. Following the membrane receptor evaluations to confirm purity, structure and bioactivity, HT-SPR can be performed with confidence to screen drug candidates. Demonstrated here are robust techniques to explore binding of drug candidates against membrane receptors using HT-SPR. These techniques highlight that with stable receptor formats, both generalized binding and detailed kinetics can be ascertained. Importantly, optimized buffer conditions must be determined for each format that stabilize the receptor and allow for confidence in the reported measures of binding. The throughput of the Carterra LSA® reduces the time the receptor must remain stable and helps to mitigate losses in activity which can occur on lower throughput screening approaches that require longer to complete a screening assay. While this work was done using intact monoclonal antibodies, the approach could easily be extended to any drug format including nanobodies, scFvs, aptamers, and even arrays of DEL compounds. The capacity allows for up to 384 drug candidates to be arrayed in the surface simultaneously and up to 1,152 candidatesscreened in a single unattended run.
This web search service is supported by Google Inc.